RNA sequencing is the molecular technique used to identify the order of nucleotide bases (adenine, uracil, guanine, and cytosine) in an RNA molecule.
RNA sequencing (RNA-Seq) is a newly developed next-generation sequencing approach to the transcriptome (a complete set of RNA content within a cell including both coding and non-coding ones) profiling. It has been very effective in transcriptomics, revealing molecular constituents of cells, studying gene expression, disease study and diagnosis, pharmaceutical developments, and ribosomal profiling.
In 1965, Robert W. Holley for the first time sequenced 77 nucleotides of a yeast tRNA. But, the modern RNA-Seq technique is developed only in 2008, shortly after the development of the next-generation sequencing (NGS) technique.
Interesting Science Videos
Principle of RNA Sequencing
Its basic is similar to gene sequencing (DNA sequencing), but the order of nucleotide sequences of a target RNA molecule is obtained instead of a DNA molecule. Initially, the Sanger Sequencing technique was used for RNA sequencing; however, shortly after the introduction of the NGS technique, a high-throughput sequencing technique came into use.
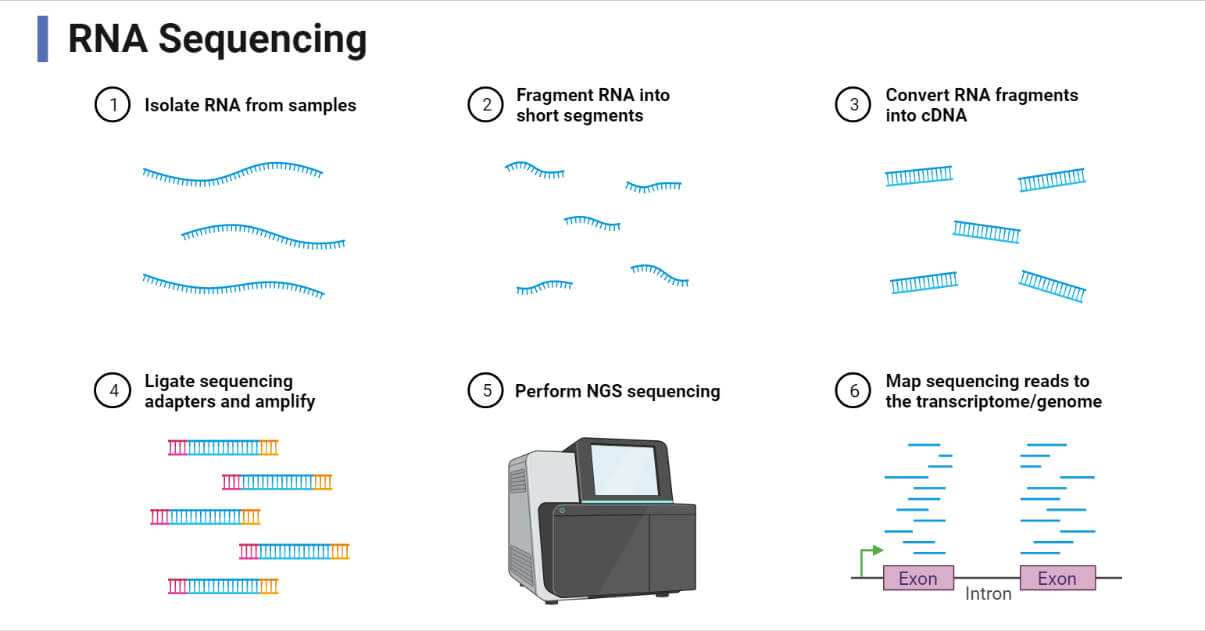
The basic protocol for RNA-Seq includes RNA extraction, isolation of target RNA and purification, cDNA synthesis, adaptor ligation, library preparation, fragmentation, and sequencing and data analysis.
Types of RNA Sequencing
On the basis of the formation of cDNA
- Direct RNA Sequencing
In this method, isolated RNAs are sequenced directly without prior converting them into cDNA. Since RNAs are unstable as compared to DNAs, they are difficult to handle and work with. However, the conversion of RNAs into cDNAs introduces various biases, error points, and disturbances that interfere with the accurate sequencing process. Additionally, cDNA preparation is a multistep complex process and cDNAs are not even suitable for sequencing smaller RNAs.
- Indirect RNA Sequencing
It is also called complementary DNA sequencing. In this type, RNAs are first converted into cDNAs and then the cDNAs are sequenced. Conversion of RNAs into cDNAs makes them more stable and easy to sequence.
On the basis of types of RNA sequenced
- Whole Transcriptome RNA-Sequencing (Total RNA-Seq)
Whole transcriptome sequencing (WTS) develops sequences of all types of RNAs present in the sample. As it profiles the entire transcriptome, it provides all the required information about gene expression and nucleotide of a cell.
- mRNA-Sequencing
In this method, only mRNAs are sequenced. mRNAs are first isolated using poly-A chromatography or poly-A magnetic beads and forming a poly-A library. The library is then sequenced either directly or indirectly to get an mRNA sequence.
- tRNA-Sequencing and rRNA-Sequencing
tRNAs are isolated and sequenced in tRNA-Seq. Similarly, rRNAs are sequenced in rRNA-Seq. Both these types are rarely used.
- Targeted RNA-Sequencing
It is the method of sequencing a specific transcript of interest.
- Small RNAs Sequencing
In this sequencing type, small non-coding RNAs of a cell are sequenced. The most commonly sequenced small RNAs are miRNA, siRNA, and piRNA.
- Single Cell RNA Sequencing
In this method, RNAs extracted from a single cell line/type are sequenced. All the transcripts of a single cell are captured, transcript libraries are developed, and the whole library is sequenced.
Procedure/Steps of RNA Sequencing
The indirect method of RNA-Sequencing i.e. cDNA formation method is the most widely used method of RNA-Seq, so in this article, we will describe a general step of the indirect RNA-Seq method. The general workflow of RNA-Seq can be summarized as:
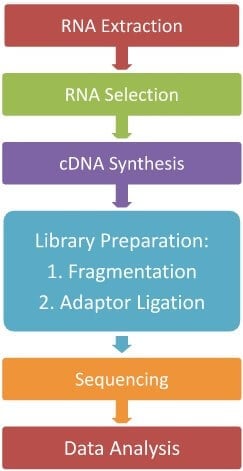
- RNA Extraction
The first step of RNA-Seq is the lysis of the cell and complete transcriptome extraction. RNA lysis buffer and organic solvent-based RNA isolation methods are widely used for cell lysis and RNA extraction. The extracted RNAs are then washed and purified as DNA-free RNAs and kept in buffer or RNase-free water.
- RNA Selection
The RNA content of a cell is huge; there are wide varieties of RNAs including the most common types; rRNA, tRNA, and mRNA. From the extracted transcriptome, certain types of RNAs we need to sequence are selected by several processes like affinity chromatography, electrophoresis, filtration (size exclusion), enzymatic depletion, target enrichment/depletion, etc. If you are planning for whole transcriptome RNA-Seq, there is no need to select specific RNA.
mRNAs are mostly selected and sequenced because they are the direct transcripts of a gene containing coding sequences. Poly-A library formation is the most common method used to isolate mRNAs from the complete set of RNAs.
- cDNA Synthesis
The isolated and selected RNAs are subjected to a reverse transcription process in order to transcribe the sample RNAs into more stable first-strand cDNAs. The first-strand cDNAs are then amplified using the Taq DNA polymerases and nucleotides forming the second-strand cDNAs.
- Selection
It is an optional process done to get rid of rRNA molecules (which occupy about 80% to 90% of total cellular RNA content), globin, and other smaller RNAs. This step will optimize the RNA sequencing process promoting efficiency of the process and saving money, time, and reagents. Target enrichment, rRNA depletion, probe-based depletion, and enzymatic depletion methods can be used for selecting the required cDNA.
- Library Preparation
cDNA library is a collection of total cDNA synthesized for sequencing. Library preparation includes:
Fragmentation and Size Selection
It is also an optional process to optimize the sequencing of targeted RNAs/cDNAs. The cDNAs are fragmented and fragments of a certain size are selected. Fragmentation can be done by chemical/enzymatic processes or physical processes like sonication.
Adaptor Ligation
At the end of fragmented and/or selected cDNAs adaptors are ligated. Adaptors are short synthetic oligonucleotides that bind transcripts and serve as a priming site for sequencing.
Indexing and Amplification
After adaptor ligation, a specific sequence (also called barcode) is added to the transcripts during PCR amplification of the cDNAs. This process is called indexing. These adaptors and barcode-ligated cDNAs are amplified to increase the concentration of the developed library.
- Sequencing
The finally prepared cDNAs in the cDNA library are sequenced using the high throughput next generation sequencing (NGS) method in the high throughput NGS machine. The process of sequencing is similar to the DNA sequencing method. Each cDNA fragment is read individually and the obtained data are analyzed using bioinformatics tools.
- Analysis of RNA Sequencing
Analyzing the sequence reads and obtaining a complete transcriptome sequence is an arduous process. In general, the obtained reads are either arranged and compared with reference sequences for testing the presence of certain genes/RNA or assembled for obtaining the complete sequence of the test RNAs.
The general workflow of RNA-Seq data analysis includes the following steps:
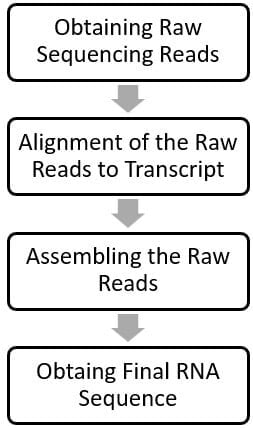
Obtaining Raw Reads
The high throughput NGS machine generates sequences of each fragment of cDNA/RNA; mostly in FASTQ-format files. These random data containing unarranged sequences and different fragments of cDNAs/RNAs are called the Raw Reads.
Alignment of the Raw Reads
The Raw Reads are fed to bioinformatics tools containing read mapping algorithms which are programmed to align the Raw Reads to the genome. The raw reads don’t contain introns, so aligning the raw reads to a genome (containing both introns and exons) is tricky and difficult; however, different software/tools like “splicing-aware” aligners are available to do the job. These tools can detect the reads across the exon-intron boundaries and align the raw reads properly leaving the intron parts. Different raw read-aligning tools available are GSNAP, MapSplice, STAR, RUM, TopHat, BaySeq, EdgeR, MISO, PEER, HCP, etc.
Assembling the Raw Reads
After aligning the raw reads, they are assembled into a transcript (sequence of RNA) using different to obtain a complete sequence of the sample RNA. It is generally done by using any of the two methods, viz. De novo Reconstruction Method and Genome Guided Method.
De novo Reconstruction Method
It is the method commonly used for obtaining the sequence of unknown or incomplete genome/RNA. Software is used to assemble the contiguous transcript sequences (contigs) into a complete RNA sequence. This method is, however, full of challenges like which reads should be arranged first in the contigs, high probability of biasedness, lack of computational efficiency, etc.
Genome Guided Method
In this method, the aligned raw reads are compared with the known sequence of RNA/Genome (reference genome) available in the reference library, and the complete sequence of the sample transcript is obtained. It is mostly used if the target transcript is already known and we are just looking for a certain known RNA sequence.
Applications of RNA Sequencing
- Detection of Mutation; Mutation in genes results in the production of the altered RNAs with altered nucleotide sequences. By studying the sequence of RNA we can easily detect any kind of mutation. Even a Single nucleotide polymorphism can be detected by this method.
- Studying gene expression
- Profiling small RNAs like microRNA (miRNA), siRNAs, piRNAs, snoRNAs, etc.
- Studying undergone alternative splicing and post-transcriptional modification
- Detection of gene fusion
- Disease diagnosis; diagnosis of genetic disorders is easy with the RNA-Seq process. The presence of RNAs that can encode any unique or altered proteins relating to disorders like cancer, tumor, auto-immune diseases, genetic disorders, etc. can be promptly diagnosed.
- Species Identification and Phylogenetic Hierarchy Development; RNA-Seq data helps to identify biomarkers and unknown species and even classify them into different strains, variants, or mutants levels; especially in studying the phylogenetics of viruses.
- Ribosomal Profiling; Ribosomal profiling is the process of determining which mRNA is being translated. This will help to determine the types and amounts of protein being synthesized.
- Studying different intrinsic cellular processes, metabolic pathways and co-expression networks, embryogenesis study, etc.
- Used in pharmaceuticals like in RNA vaccinology.
Advantages of RNA Sequencing
- Have a border dynamic range than the conventional probe method and microarray method used in transcriptome analysis.
- Give more accurate measurement of gene expression than studying genome or proteins.
- It doesn’t require a probe; hence, it can be used to analyze both known and unknown transcripts.
- It can be used to analyze the whole transcriptome and get both qualitative as well as quantitative results.
Limitations of RNA Sequencing
- It is expensive and time-consuming – requiring time-intensive processes like sample preparation, assay designing, library preparation, running assay, and analyzing data.
- Demand high throughput and sensitive targeted sequencing approach which are not yet possible to get practically.
- Lack of optimized and hard-and-fast protocol of RNA sequencing.
- Analyzing a single type of RNA is very difficult because a cell contains numerous types of RNAs which are difficult to classify and isolate.
- No strict raw reads assembling method for unknown or novel transcript indicates biasedness or error in obtained sequencing result.
Common Examples of RNA Sequencing Platforms Used in RNA-Seq
- Illumina HiSeq Platform
- Illumina NovaSeq X and NocaSeq X Plus Sequencing System
- MACE-Seq-3’mRNA/UTR Sequencing Platform (GeneXPro Company)
- PacBio Single-Molecule Real-Time (SMRT) Sequencing Platform
- Luminex xMAP INTELLIFLEX Multiplexing System
References
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017 Feb 16;65(4):631-643.e4. doi: 10.1016/j.molcel.2017.01.023. PMID: 28212749.
- Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018 Aug 7;50(8):1-14. doi: 10.1038/s12276-018-0071-8. Erratum in: Exp Mol Med. 2021 May;53(5):1005. PMID: 30089861; PMCID: PMC6082860.
- Kukurba KR, Montgomery SB. RNA Sequencing and Analysis. Cold Spring Harb Protoc. 2015 Apr 13;2015(11):951-69. doi: 10.1101/pdb.top084970. PMID: 25870306; PMCID: PMC4863231.
- Lucchinetti E, Zaugg M. RNA Sequencing. Anesthesiology. 2020 Nov 1;133(5):976-978. doi: 10.1097/ALN.0000000000003524. PMID: 32833386.
- Wang, Zhong; Gerstein, Mark; Snyder, Michael (2009). RNA-Seq: a revolutionary tool for transcriptomics. , 10(1), 57–63. doi:10.1038/nrg2484
- Ziegenhain et al., 2017, Molecular Cell 65, 631–643 February 16, 2017 ª 2017 Elsevier Inc. http://dx.doi.org/10.1016/j.molcel.2017.01.023
- Koch, C. M., Chiu, S. F., Akbarpour, M., Bharat, A., Ridge, K. M., Bartom, E. T., & Winter, D. R. (2018). A Beginner’s Guide to Analysis of RNA Sequencing Data. American Journal of Respiratory Cell and Molecular Biology, 59(2), 145-157. https://doi.org/10.1165/rcmb.2017-0430TR
- RNA Sequencing- Principle, Steps, Methods And Applications (geneticeducation.co.in)
- What is RNA sequencing? – YourGenome
- RNA-Seq: Basics, Applications and Protocol | Technology Networks
- A Brief History of RNA-Seq | RNA-Seq Blog
- Ozsolak, F., Platt, A., Jones, D. et al. Direct RNA sequencing. Nature 461, 814–818 (2009). https://doi.org/10.1038/nature08390
- Types of DNA and RNA Sequencing | IDT (idtdna.com)
- RNA Sequencing | RNA-Seq methods & workflows (illumina.com)
- What is RNA Seq and Why Use RNA Sequencing? – Zymo Research International
- Limitations of RNA-Seq — BioSpyder
