Reverse vaccinology is a new technique in vaccine development in which computational technologies are used to find possible vaccine targets from pathogen genomes.
This technique has transformed vaccine design, resulting in the discovery of potent vaccinations against a wide spectrum of infectious diseases. We shall look into reverse vaccinology in this essay, including its history, principles, methodology, and applications.
Interesting Science Videos
Principles of Reverse Vaccinology
It entails identifying possible vaccine targets from pathogen genomes using computational methods. The main premise is to discover proteins that are required for the pathogen’s survival or virulence and are also present on the pathogen’s surface. Vaccines can then target these proteins to elicit an immune response that will protect against illness.
History of Reverse Vaccinology
- Vaccination is an ancient medical practice with the first reports of using this method of treatment from Asian regions as lesions of smallpox were used as injectable material to protect against infections and serious diseases.
- Then, Edward Jenner, in the year 1796, coined the term ‘Vaccine’ for the injectable material from cows and transmitting it to individuals affected by smallpox.
- Louis Pasteur, in his discovery of infections caused by microorganisms in the 1800s, laid the foundation for a rational vaccine development process and a basic rule to be followed for vaccinology, which states ‘isolate, inactivate and inject the microorganism’ for making a vaccine against the disease caused.
- By the end of the 20th century, all vaccines were developed by following the Pasteur rule of vaccinology.
- But with the development of new technologies, such as the chemical conjugation of proteins to other biomolecules like polysaccharides, and Recombinant DNA, vaccine development progressed further.
- And when Craig Venter deduced the ‘genome’ of the first free-living organism, which revolutionized the vaccine development progression.
- With the aid of computational tools, scientists were able to design vaccines by analyzing the specific microorganism’s genome without the need for growth in lab culture media.
- The first pathogen analyzed for the reverse vaccinology method was ‘Men B’ (meningococcus), a bacterial pathogen responsible for 50% of meningococcal meningitis worldwide.
- Vaccines against Streptococcus bacteria devised via reverse vaccinology contain four proteins that can provide immunization against the pathogen for all blood types.
And as this approach is gaining more consideration, more and more vaccines are devised against harmful pathogens. This approach can also help in fighting against the fast evolutionary nature of pathogen drug resistance.
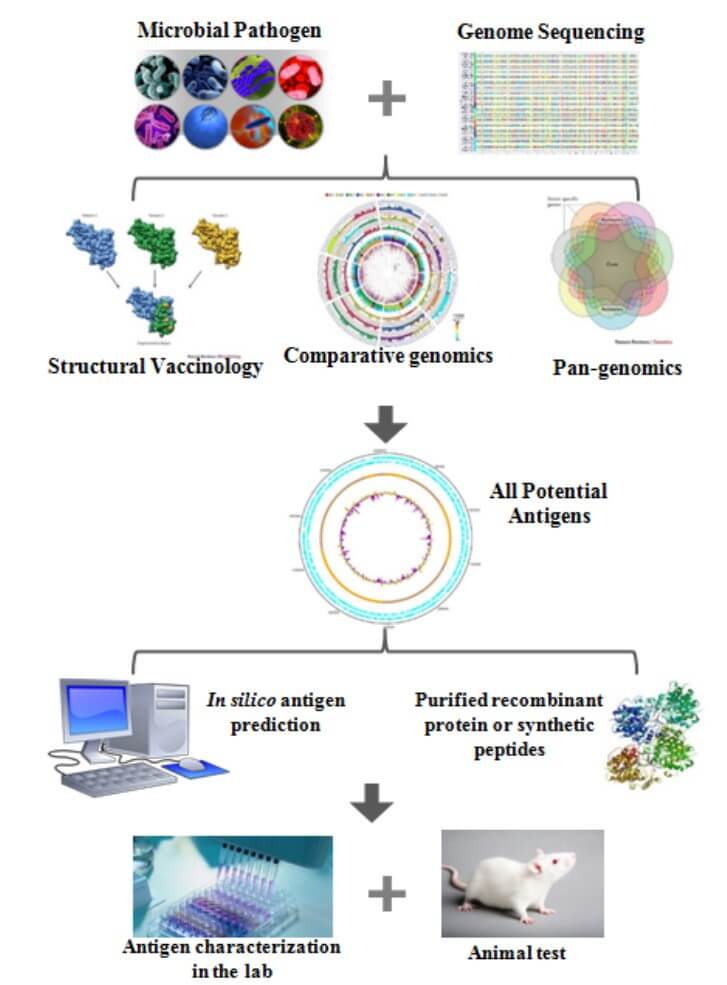
Methods and Application
Reverse vaccinology is a modern approach to vaccine development that involves the use of computational tools to identify potential vaccine targets from pathogen genomes. This technique has revolutionized the way vaccines are designed and has led to the development of effective vaccines against a wide range of infectious diseases. Keep looking for details, and we will explore the methods and applications of reverse vaccinology.
Methods of Reverse Vaccinology
The methods used in reverse vaccinology typically involve a combination of bioinformatics, genomics, proteomics, and immunology. The process typically involves the following steps:
- Sequencing the Pathogen’s Genome: The first stage in reverse vaccinology is to sequence the pathogen’s genome. This is possible with next-generation sequencing methods.
- Analysis of the Genome Sequence: The genome sequence is then studied using bioinformatics techniques to find genes involved in virulence, pathogenesis, or other key biological processes. Various methods and software tools are used in this step to predict protein localization, identify domains, and assess possible immunogenicity.
- Proteomics Analysis: The selected genes are then expressed, and the resulting proteins are examined using proteomics techniques to identify proteins on the pathogen’s surface. During this step, tools such as mass spectrometry, 2D electrophoresis, and protein microarrays are used.
- Immunological Validation: The identified proteins are then tested for their ability to stimulate an immune response in animals or humans. This typically involves the use of animal models or in vitro assays such as ELISA, Western blotting, and flow cytometry. This step aims to confirm the immunogenicity and protective potential of the identified targets.
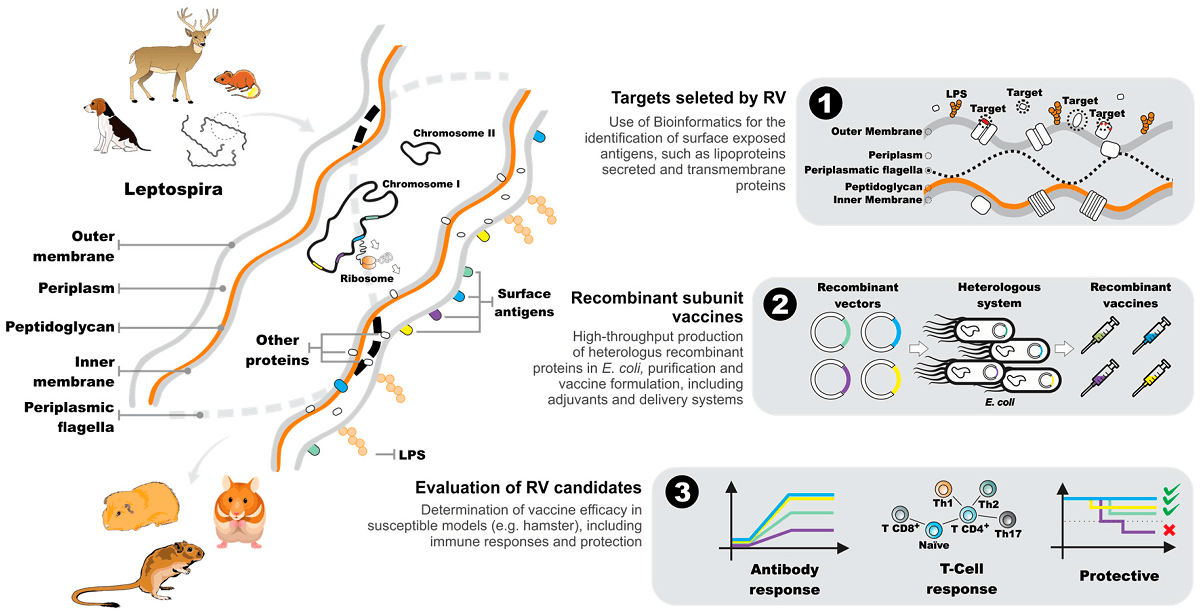
Applications of Reverse Vaccinology
Reverse vaccinology has been used to develop vaccines against a wide range of infectious diseases, including bacterial, viral, and parasitic infections. Some examples of vaccines developed using reverse vaccinology include:
- Bexsero is a vaccine that protects against Neisseria meningitidis. Bexsero was the first vaccine designed utilizing reverse vaccinology, and it targets the bacterium’s surface-exposed proteins.
- Prevnar 13 is a Streptococcus pneumonia vaccination. Prevnar 13 targets proteins involved in the development of bacterial capsules, which is a major pathogenicity factor.
- Shingrix is a herpes zoster vaccination. Shingrix targets glycoproteins on the virus’s surface.
- Mosquirix is a malaria vaccine that protects against Plasmodium falciparum, the parasite that causes malaria. Mosquirix targets surface-exposed proteins in the parasite’s sporozoite stage.
- Vaccines against emerging infectious illnesses, such as COVID-19, have also been developed using reverse vaccinology. The technique enabled researchers to quickly discover possible vaccine targets for the SARS-CoV-2 virus, leading to the development of numerous successful vaccines, including the Pfizer-BioNTech and Moderna vaccines.
Future Aspects of Reverse Vaccinology
It provides a number of advantages over standard vaccine development methods. It enables more rational and efficient identification of prospective vaccination targets and has the potential to speed up the vaccine development process. It can also discover targets that are conserved across different pathogen strains or serotypes, paving the way for the development of broad-spectrum vaccines. Furthermore, reverse vaccinology can reveal targets that are less immunogenic, allowing for the development of vaccinations that are more efficacious and have fewer adverse effects.
Personalized Vaccine Development
- It has the potential to be utilized in the development of personalized vaccines. vaccinations that are tailored to an individual’s specific genetic composition, immunological response, and environmental exposures are known as personalized vaccinations.
- Reverse vaccinology provides the ability to develop vaccine targets tailored to a person’s disease exposure and immune response.
- This could lead to the development of more effective vaccines with fewer adverse effects. Individualized vaccines could be especially beneficial for those who are at high risk of contracting specific illnesses, such as those with compromised immune systems or a history of recurring infections.
- Personalized vaccines could also generate vaccinations for rare or emerging infectious illnesses when typical vaccine development methods may be impractical.
Limitations of Reverse Vaccinology
However, there are significant obstacles to developing tailored vaccinations utilizing reverse vaccinology.
- The requirement for enormous amounts of genetic and proteomic data from individuals to find customized vaccination targets is one obstacle.
- Another barrier is the high cost and logistics of mass-producing customized vaccinations. Despite these obstacles, the potential benefits of tailored vaccines make this a critical area of research in reverse vaccinology.
- Personalized vaccinations are likely to become a significant tool for preventing and treating infectious diseases as our understanding of the immune system and pathogen-host interactions grows.
Traditional vaccinology vs. Reverse vaccinology
| Characters | Traditional Vaccinology | Reverse Vaccinology |
|---|---|---|
| Antigens available | Using biochemical and genetic tools, only a limited number of antigens can be used. | All antigens can be identified using genetic tools |
| Property of antigens | The most abundant and immunogenic antigens of cultivable microbes can be identified. | All antigens of all types of microbes can be identified. |
| Immunology of the antigens | Variable antigens, that elicit a greater immune response. Since some mimic self-antigens, thus cause autoimmunity. | Conserved antigens can be detected, which are not very immunogenic. Self-antigens can be negatively selected. |
| Polysaccharide antigens | Important vaccine candidate | Cannot be used in reverse vaccinology. To discover carbohydrate antigens, operons coding for polysaccharides can be detected. |
| T cell epitopes | Known epitopes are limited to the known antigens. | Virtually every single T cell epitope is available. Screening of the total T cell immunity can be done by overlapping peptides. |
Conclusion
To summarise, reverse vaccinology is an effective vaccine development strategy that employs computational techniques, genomes, proteomics, and immunology to find possible vaccine targets. This technique has transformed vaccine design, resulting in the discovery of potent vaccinations against a wide spectrum of infectious diseases. Reverse vaccinology has various advantages over traditional vaccine development methods, including the capacity to find targets that are conserved across different pathogen strains or serotypes, allowing for the production of broad-spectrum vaccines. Furthermore, it has the potential to be used for tailored vaccines, which might be especially beneficial for people who are at high risk of contracting specific infections. While there are still challenges associated with the development of personalized vaccines using reverse vaccinology, the potential benefits make this an important area of research in the field of vaccinology.
References
- Rappuoli, Rino. “Reverse vaccinology.” Current opinion in microbiology 3.5 (2000): 445-450.
- Rappuoli, Rino, et al. “Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design.” Journal of Experimental Medicine 213.4 (2016): 469-481.
- Sahin, Ugur, and Özlem Türeci. “Personalized vaccines for cancer immunotherapy.” Science 359.6382 (2018): 1355-1360.
- Sette, Alessandro, and Rino Rappuoli. “Reverse vaccinology: developing vaccines in the era of genomics.” Immunity 33.4 (2010): 530-541.
- Personalised Vaccines: A New Era in Cancer Immunotherapy – Schumacher and Schreiber (2015)
