Proteomics is the study of the proteome, which is the complete set of proteins that are produced in a biological system.
Proteomics provides direct insights into cellular functions because proteins carry out most of the gene functions in cells. Proteomics focuses on the study of proteins, their structures, interactions, and functions.
Proteomics can be used to study the expression of proteins. Additionally, proteomics can reveal how proteins are modified, such as by post-translational modifications. Proteomics can also provide information on the movement of proteins between different subcellular compartments, as well as their involvement in metabolic pathways and interactions with other proteins.
Interesting Science Videos
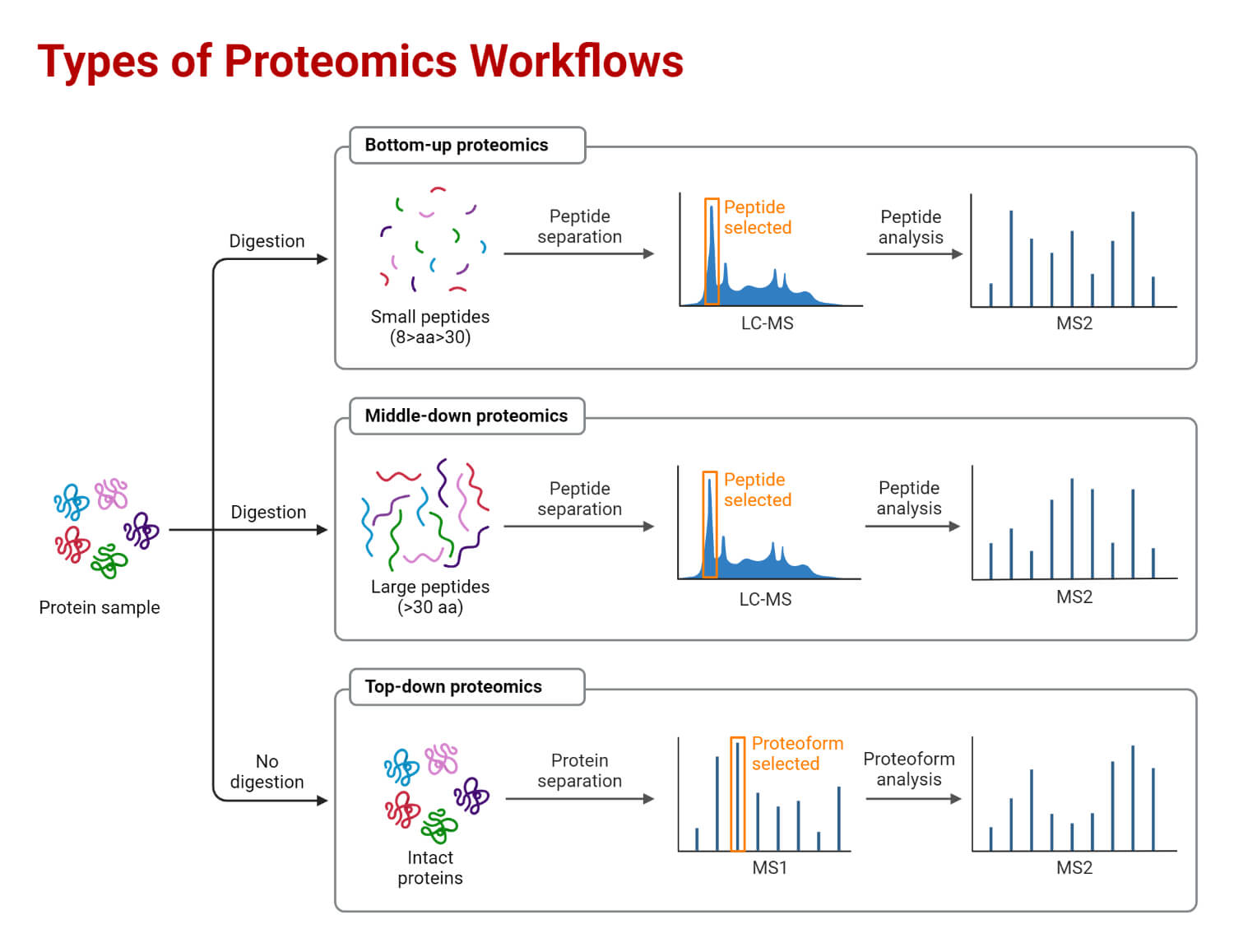
Types of Proteomics
Proteomics can be classified into three categories; expression proteomics, structural proteomics, and functional proteomics.
- Expression proteomics is a field that studies the changes in protein expression, both qualitatively and quantitatively, under different conditions. It detects variations in protein expression in different cells, such as tumor tissue compared to normal tissue. Expression proteomics experiments often use techniques such as 2D gel electrophoresis and mass spectrometry to detect and quantify proteins.
- Structural proteomics is concerned with determining the three-dimensional structure of proteins. It uses techniques such as X-ray crystallography and nuclear magnetic resonance spectroscopy (NMR) to determine the precise arrangement of atoms within a protein. Structural proteomics can also be used to investigate protein-protein interactions and the interactions between proteins and other biomolecules such as DNA or RNA.
- Functional proteomics is focused on understanding the functions of proteins and their interactions with other molecules in the cell. It involves identifying the protein complexes and interactions that are involved in specific biological processes, as well as the roles of individual proteins within these complexes.
Proteomics Methods and Steps
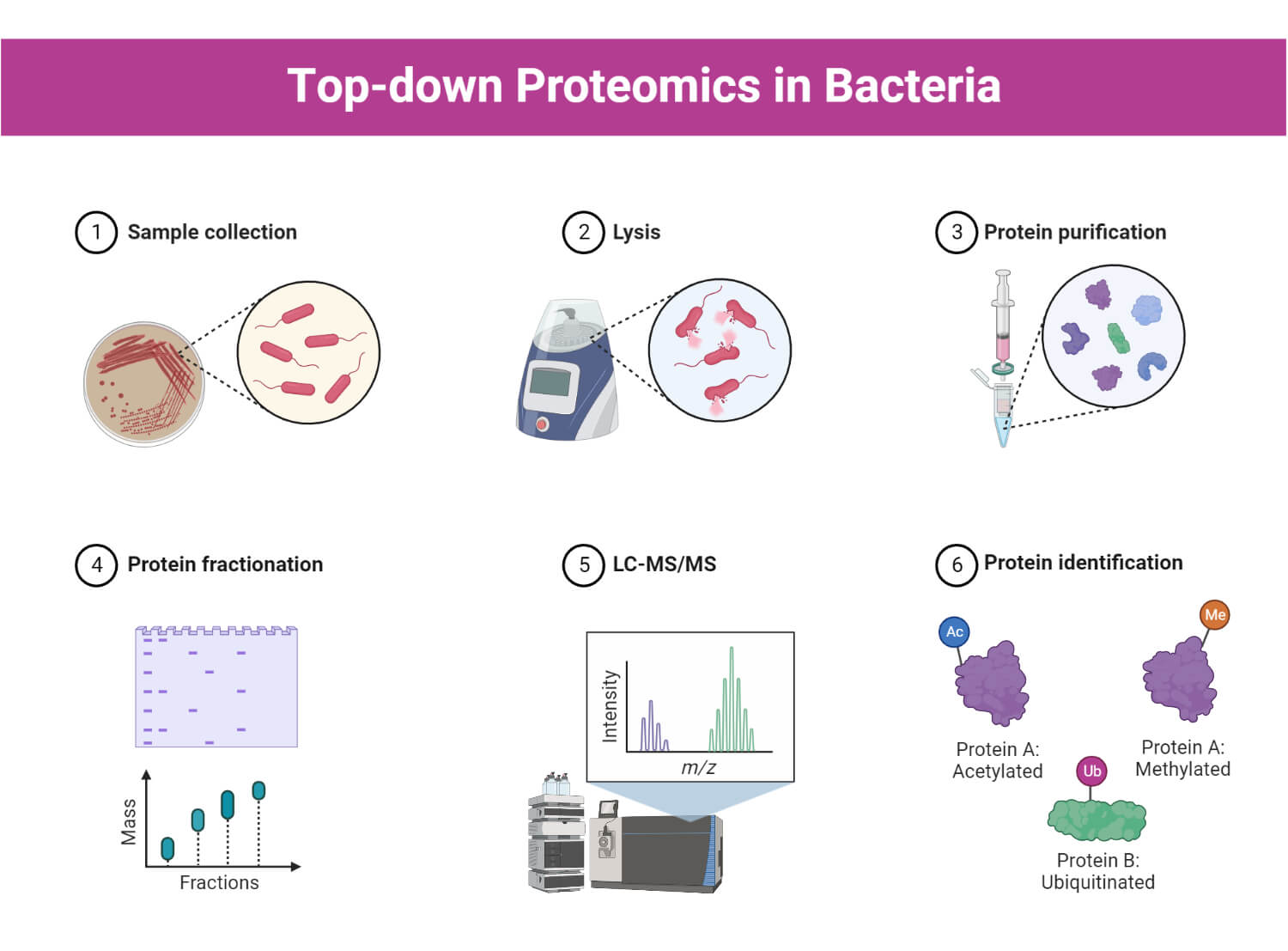
Proteomics uses several high-throughput technologies to investigate and analyze proteomes. Proteomic procedures consist of several steps, including sample preparation, separation of proteins, protein identification, and validation.
1. Sample preparation
- Sample preparation is a critical step in proteomics experiments, which involves the extraction and purification of proteins from biological samples for further analysis.
- Generally, the extraction of proteins from a mixture involves a combination of chemical and physical methods.
- Organic solvents and detergents are commonly used in sample preparation to enhance protein solubility and disrupt cellular membranes to release intracellular proteins.
- Tissue disruption techniques, such as mechanical homogenization or sonication, are often used in sample preparation to break down tissues and release proteins.
2. Separation and isolation of proteins
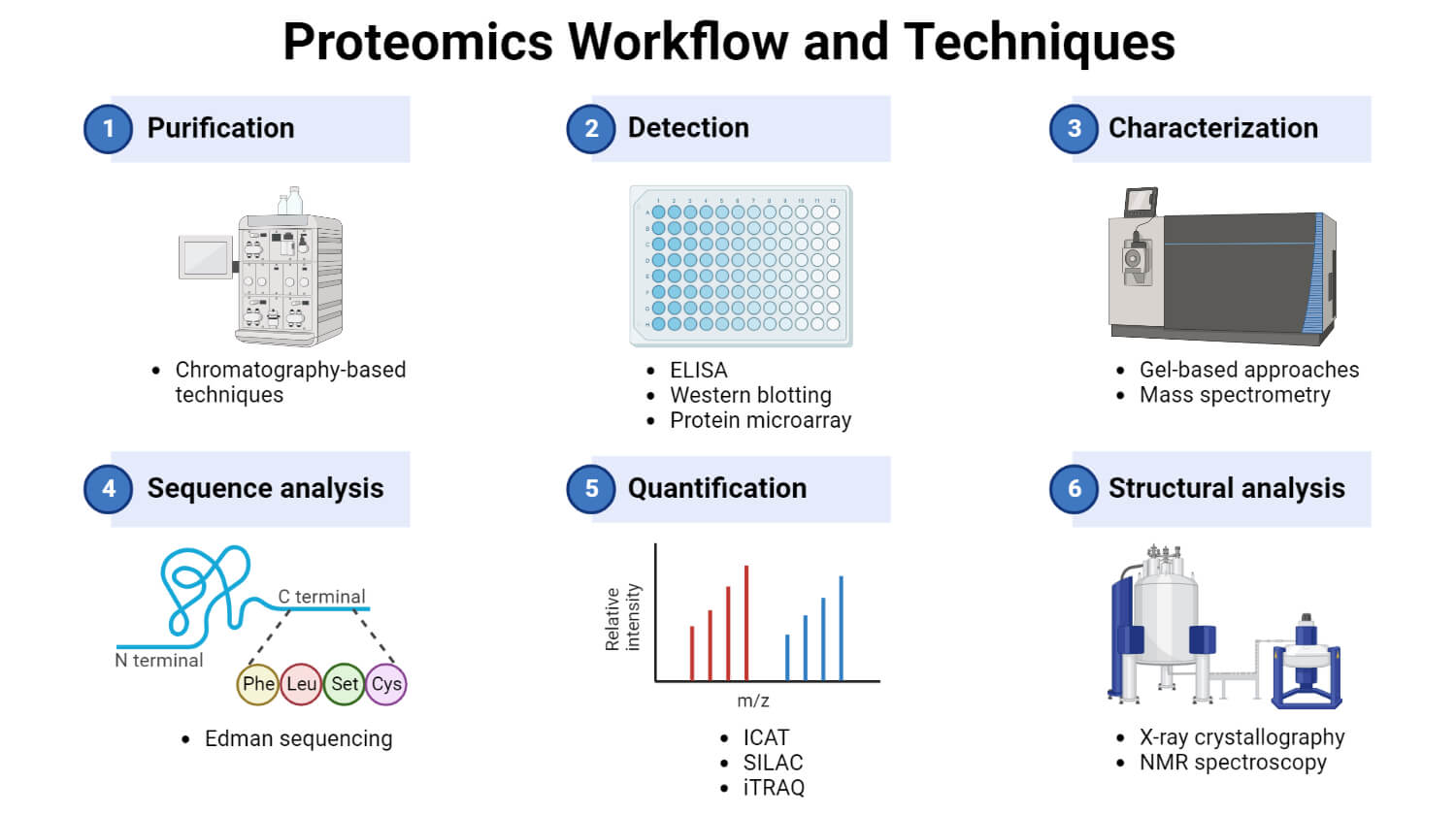
Two approaches are mainly used for the separation of proteins: gel-based and chromatography-based.
Gel-based approach
1-DE
- One-dimensional gel electrophoresis (1-DE), also known as SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), is a commonly used method for separating proteins based on their molecular weight.
- SDS is a detergent used to denature proteins and impart a uniform negative charge proportional to their mass. This uniform negative charge allows the separation of proteins based on size during electrophoresis.
- The SDS-treated proteins are then separated in a polyacrylamide gel matrix using an electric field. The smaller proteins move more quickly through the gel, while the larger proteins move at a slower pace.
2-DE
- Two-dimensional gel electrophoresis (2-DE) is a technique used to separate proteins on the basis of two parameters: isoelectric point and molecular weight.
- In the first separation step, called isoelectric focusing (IEF), proteins are separated on the basis of their isoelectric point (pI), which is the pH at which the protein has no net charge.
- The second separation step is SDS-PAGE, where proteins are separated based on their molecular weight.
- This separation by two different parameters allows for better differentiation of proteins and can also detect post-translational modifications and different forms of proteins that cannot be resolved by 1-DE.
- However, 2-DE has limitations, as it cannot detect proteins with low molecular weight or those that are too large.
Chromatography-based approach
- IEC (Ion Exchange Chromatography) is a chromatographic technique used to purify proteins based on their charges. It works by using a charged stationary phase that interacts with oppositely charged proteins present in the sample.
- SEC (Size exclusion chromatography) separates molecules based on their size. It uses a porous resin as the stationary phase and molecules that are larger in size are prevented from entering the pores, while smaller molecules can penetrate the pores and take a longer time to elute.
- Affinity chromatography separates proteins based on their interaction with an immobilized ligand. This method is useful in reducing the complexity of proteins in 2-DE and non-2-DE experiments.
- LC (Liquid chromatography) is one of the most widely used methods in proteomics to separate proteins from complex mixtures. It is a valuable tool in proteomics research that can be used to analyze large and fragile biomolecules.
3. Protein identification and characterization
Mass spectrometry
- Mass spectrometry (MS) is a powerful tool that can rapidly sequence proteins and determine their molecular weights.
- It works by ionizing protein molecules and then calculating their mass based on mass-to-charge ratios.
- Two methods used for ionization in MS are ESI (Electrospray Ionization) and MALDI (Matrix-Assisted Laser Desorption Ionization).
- In ESI, the protein sample is activated to create charged droplets that increase gaseous ion production. The ions are then analyzed with a mass analyzer.
- In MALDI, a chemical matrix is mixed with the peptides and spotted onto a metal plate to make a crystal lattice. The matrix absorbs energy and transfers it to the sample. Then the peptide ions are detected by a mass analyzer.
Database searching
- Once peptides have been sequenced, bioinformatics programs can be used to search for the identity of a protein in a database of known proteins.
- The aim of the search is to find exact or nearly exact matches. The user must provide as much information as possible, such as molecular masses of peptide, peptide sequence, molecular weight, and isoelectric point of the intact protein, to obtain a unique identification of a particular protein.
- This method is limited to model organisms that have completely sequenced and well-annotated genomes.
Differential In-Gel Electrophoresis (DIGE)
- It is a method used to detect differences in protein expression patterns between two samples.
- It involves labeling proteins from two different samples with different fluorescent dyes and mixing them together before separating them by electrophoresis on a two-dimensional gel.
- The differentially expressed proteins in both samples can then be visualized and compared on the same gel.
- However, there are some limitations to this approach, such as, different proteins may take up fluorescent tags to different extents, and some proteins labeled with fluorophores may become less soluble and precipitate before electrophoresis.
Protein microarrays
- Protein microarrays are high-throughput devices that contain high-density grids with immobilized proteins.
- Protein microarrays were first developed as immunoassays or antibody arrays which are a type of microarray that involves capturing proteins on an antibody microarray substrate and detecting them using antibodies.
- A new technique uses protein scaffolds that are smaller, more stable, and more specific than antibodies to capture target proteins.
- Another technology under development is Protein-Print, which uses molecular imprinting to capture molecules with high specificity.
Applications
Proteomics has a wide range of applications in various fields. Some of these applications are:
- Proteomics can be used in biomarker discovery to identify protein markers for the diagnosis and prognosis of different diseases.
- Proteomics can also be used to identify potential drug targets and develop more effective therapies by studying the protein-protein interactions in diseased cells.
- Proteomics is used in agriculture to study plant-pathogen interactions and develop crops that are more resilient to environmental stresses.
- Proteomics is also used in the field of food science to improve the nutritional value and safety of foods and to detect allergens.
- Proteomics can be used to identify proteins that are associated with drug toxicity, leading to the development of safer drugs.
- Proteomics also helps in the development of personalized medicine, where the proteome of individual patients can be analyzed to tailor treatments based on their specific genetic makeup.
References
- Al-Amrani, S., Al-Jabri, Z., Al-Zaabi, A., Alshekaili, J., & Al-Khabori, M. (2021). Proteomics: Concepts and applications in human medicine. World Journal of Biological Chemistry, 12(5), 57-69. https://doi.org/10.4331/wjbc.v12.i5.57
- Graves, P. R., & J. Haystead, T. A. (2002). Molecular Biologist’s Guide to Proteomics. Microbiology and Molecular Biology Reviews, 66(1), 39-63. https://doi.org/10.1128/MMBR.66.1.39-63.2002
- https://www.ebi.ac.uk/training/online/courses/proteomics-an-introduction/what-is-proteomics/
- https://www.technologynetworks.com/proteomics/articles/proteomics-principles-techniques-and-applications-343804
- Joshi, K., & Patil, D. (2017). Proteomics. Innovative Approaches in Drug Discovery, 273–294. doi:10.1016/b978-0-12-801814-9.00009-x
- Liang, K.-H. (2013). Proteomics. Bioinformatics for Biomedical Science and Clinical Applications, 83–106. doi:10.1533/9781908818232.83
- Twyman, R. M. (2012). Proteomics. Encyclopedia of Applied Ethics, 642–649. doi:10.1016/b978-0-12-373932-2.00047-8
- Xiong, J. (2006). Essential Bioinformatics. Cambridge: Cambridge University Press. doi:10.1017/CBO9780511806087
