Introns were discovered in 1977 when DNA sequencing was introduced. Introns were found in the protein-coding genes of adenovirus and then found in t-RNA, and ribosomal RNA genes. Introns were introduced by Phillip Allen Sharp and Richard J. Roberts. The term Introns was introduced by Walter Gilbert.
What are Introns?
Introns are non-coding regions in DNA found between exons in a gene. Introns in genes do not code for amino acids. In cells, a portion of the gene sequence that is not expressed for proteins is called introns. In eukaryotic hnRNA introns are common, but in prokaryotes, they are present in tRNA and rRNA. The length of introns differs among species and genes in the same species; mammals and flowering plants have multiple introns and are longer than exons. Some organisms like Saccharomyces cerevisiae and protists have fewer introns. Introns can also do alternate splicing of a gene because many different proteins have some common sequences so, it can be translated from a single gene.
Structure of Introns
These introns are longer than exons and consume 90% of the gene; introns are general in genes; human genes contain 90 percent of introns, an average of nine introns per gene.
A stretch of DNA called introns starts and ends with a particular nucleotide sequence; these sequences represent a boundary between introns and exons, which is knowns as a splice site. The boundary recognition between coding and non-coding DNA is essential for creating functioning genes. In humans and vertebrates, introns start with 5′ GUA and end with CAG 3′; there are also conserved sequences found in vertebrates and invertebrates having a branch in a loop formation.
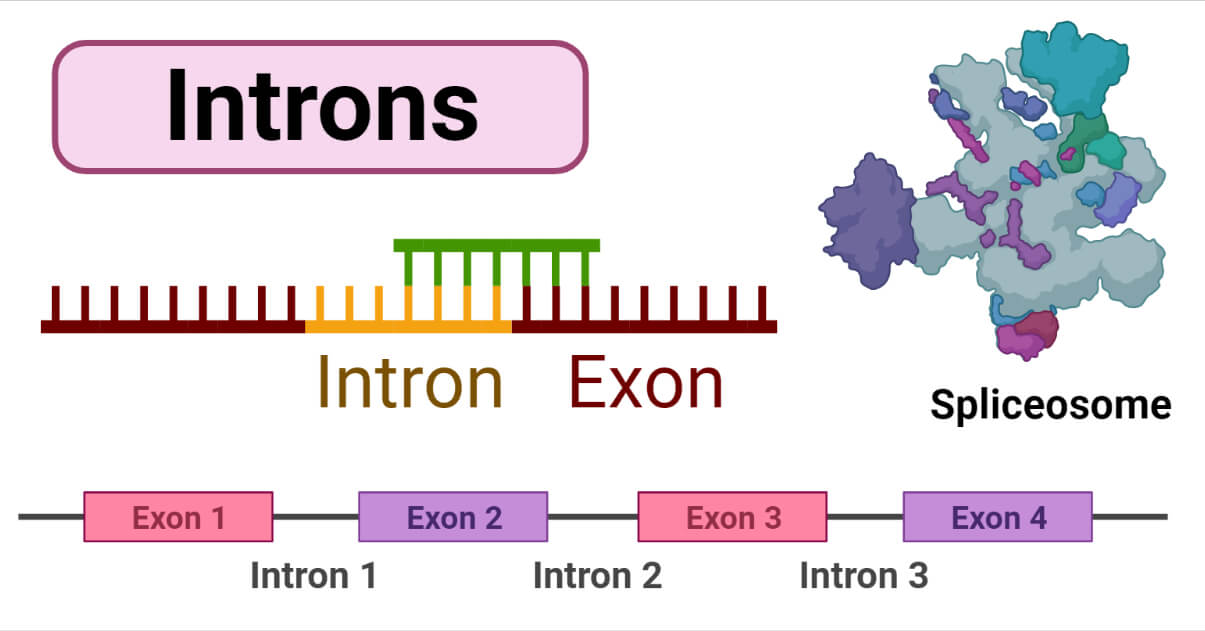
Introns Functions
As introns are considered junk DNA, It has a role in regulation and gene expression.
Introns are responsible for the increase in gene length. This increases the possibility of crossing over and recombination between sister chromosomes.
An increase in genetic variation shows new gene variants by duplication, deletion, and exon shuffling.
Introns also do alternative splicing means a single gene can encode multiple proteins.
What is Splicing?
It’s a process of removal of introns from pre-mRNA by the spliceosome. when the introns are not removed the RNA gets translated into a nonfunctional protein. So, in splicing the transcription RNA polymerase copies the complete gene including exons and introns into pre-mRNA. Before translation introns should be removed. Introns have sequences that are involved in splicing having a spliceosome recognition site, at this site spliceosomes recognize the boundary between introns and exons, and snRNPs (small nucleolar ribonucleoproteins) recognize these sites; there are many snRNPs in mRNA splicing which combined to form spliceosome.
Steps of Splicing
- The phosphodiester bond cleaves the bond between exons at 5’GU and the end of introns. There is one complimentary sequence of snRNPs (U1) at the 5′ splice site that binds to start splicing.
- Now the loop structure formation, in this, a free 5′ end combines with the branch site and conserves the sequence at the 3′ end. To initiate the loop formation, an snRNP (U2) binds to the branch site and attracts U1, then loop formation is done by a phosphodiester bond between 5′ G and A at the branch site.
- The phosphodiester cleaves the bond between the second exon and the AG 3′ end of the intron.
Introns are thousands of base pairs long and have many different cryptic splice sites which have sequence recognition available.
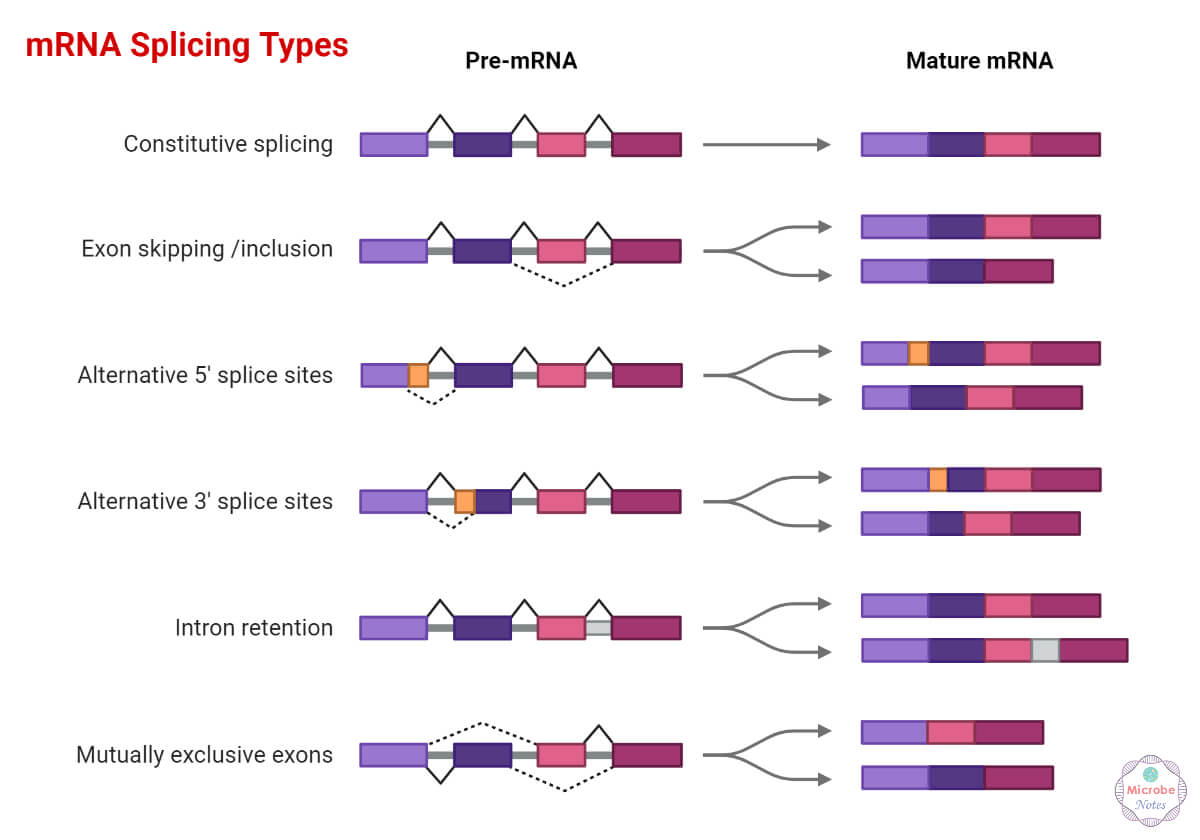
Alternative splicing
In this, introns and splicing action permits for alternative gene products or can be defined as giving different combinations of exons to get included in the final step ( mature mRNA) and making different forms of a protein called isoforms encoded by the same gene. This splicing can result in two hundred or more different mRNAs; for example, in an alternative splicing Dscam gene in Drosophila melanogaster here, a single gene has 116 exons; some are present and some are not included, so there are 18,000 different kinds of proteins found in a single gene.
Ways of alternative splicing:-
- Exon skipping- exons are not involved in final mRNA.
- Intron retention- the intron part does not get spliced correctly and stays in the final mRNA.
- Alternative splice site- spliceosome removes one or more exons and also introns.
Classification of Introns
There are Group I and Group II are ribozymes capable to catalyze their splicing out primary RNA transcript. Thomas Cech found self-splicing activity. There are four classes of introns
1) Nuclear introns
2) Group I introns
3) Group II introns
4) Transfer RNA introns
1) Nuclear introns- they are present in the nucleus of that gene (protein-coding gene) which is removed by spliceosome. For its removal, the presence of snRNAs and other proteins is important and they have branch points.
2) Group I introns- they are found in some rRNA genes. They can catalyze their self-excision from mRNA, tRNA, and rRNA precursors because they are self-splicing ribozymes. This folds into a nine-loop stem that is needed for splicing.
Group I introns are present in:-
- Mitochondria and plastids genome of plant and protists.
- The nucleus of some fungi and lichens. (rRNA genes)
- Eubacteria (t-RNA genes) and phages.
- Metazoans – in the mitochondrial gene of anthozoans.
3) Group II introns- are also present in mRNA, tRNA, and rRNA. They are self-splicing ribozymes, but the mechanism of folding is different as they fold and form a lariat structure; it is that structure that forms when introns fold back by themselves after the exon is cut.
- Highly present in the organelle genome of plants and lower eukaryotes.
- Bacterial one-quarter genome contains group II introns.
- Present in archaebacteria also.
4) Transfer RNA introns- they are found in tRNA genes and need some enzymes to splice out of genes.
| S.N. | Introns | Location | Splicing site |
| 1 | Group I | rRNA genes, organelle RNA, and some bacterial RNAs | Self-splicing (transposase) |
| 2 | Group II | Chloroplast, mitochondrial gene, prokaryotic RNAs | Self-splicing Reverse transcriptase and ribozymes |
| 3 | Nuclear | Nucleus | Non-self splicing sn RNAs |
| 4 | t-RNA | t RNA | Enzymatic |
Evolution of introns
The two theories of the introns-
IE model was given by Walter Gilbert that introns are old and numerously present in prokaryotes and eukaryotes. The introns got lost from prokaryotic organisms and were allowed to gain growth efficiency. The theory prediction was that introns were mediators that facilitated the recombination of exons such, model led to the evolution of new genes.
IL model says introns were added to original intron- less adjacent genes after deviation of eukaryotes and prokaryotes.
In this introns had their origin in parasitic transposable elements the observation made by this model was spliceosomal introns were limited to eukaryotes only.
Factors that affect the gain or loss of introns
1. organism-specific factor
- Generation time
- r-k selection strategy
- population size
- single cell or multi-cellularity
2. Gene-specific factor
- Expression level
- Alternative splicing
- Activity in time and space
- Gene function
3. Intron-specific factor
- Position
- Presence of functional elements
The similarity between introns and exons
- Have nucleic acid of a polynucleotide chain.
- Introns structure is more complex than exons.
- Introns are rich in GC region and repetitive sequence.
- Both take part in protein formation.
- Both pursue DNA packaging mechanisms to fit in the cell.
- Interact with histone protein.
- Nucleosome assembly formation, chromatin, and chromosome.
- Both are present in chromosomes.
Exons vs Introns
- Exons are a coding portion of the genome, introns are a non-coding portion of the genome.
- Exons make proteins, introns regulate gene expression.
- The final product of the exon is protein, and it is present in DNA and mRNA, but introns are present only in DNA and primary mRNA transcript not in final mRNA.
- Introns are present on heterochromatin, while exons are present in euchromatin.
References
- https://geneticeducation.co.in/introduction-to-exon-and-intron/
- https://www.bionity.com/en/encyclopedia/Intron.html
- https://www.slideshare.net/bhagatyogesh12/introns-structure-and-functions
- https://www.aatbio.com/resources/faq-frequently-asked-questions/What-are-the-types-of-introns
- https://en.wikibooks.org/wiki/Structural_Biochemistry/Nucleic_Acid/RNA/RNA_modification/Introns
- https://bio.libretexts.org/Learning_Objects/Worksheets/Biology_Tutorials/mRNA_Splicing
- https://biologydictionary.net/intron/
- https://www.nature.com/scitable/definition/intron-introns-67/
- https://www.genome.gov/genetics-glossary/Intron
