Ideonella sakaiensis is a novel bacterial species renowned for its extraordinary capacity to break down and metabolize a popular kind of plastic used in the manufacture of packaging materials, polyethylene terephthalate (PET).
Discovered recently in 2016 by a group of research scientists in Sakai city of Japan while researching the microbiome of PET-contaminated sediments at a plastic bottle recycling center, Ideonella sakaiensis has been renowned for its ability to degrade plastics. However, there is no more known about its biochemical characters, genetics, and other characteristics. But, this discovery has increased interest in the study of plastic breakdown and the potential for creating more environmentally friendly methods of handling plastic trash.
Interesting Science Videos
Classification of Ideonella sakaiensis
| Domain | Bacteria |
| Phylum | Proteobacteria |
| Class | Gammaproteobacteria |
| Order | Pseudomonadales |
| Family | Comamonadaceae |
| Genus | Ideonella |
| Species | I. sakaiensis |
Discovery of Ideonella sakaiensis
- Long before I. sakaiensis was discovered in 2016, the genus Ideonella was established in 1995 under the proposal of Malmqvist A, Welander T, Moore E, Ternstrom A, Molin G, Stenstrom I. (Reference: https://lpsn.dsmz.de/genus/ideonella)
- I. sakaiensis was discovered by a group of researchers led by Professor Kohei Oda and Professor Kenji Miyamoto of Kyoto Institute of Technology and Keio University respectively. They were researching the microbial community of PET-contaminated sediments of a plastic bottle recycling facility in the Sakai City of Japan. During their study, they discovered that about 3/4th of the PET was degraded to CO2. On further study, they found that a novel bacterium, now named Ideonella sakaiensis, first degrade and assimilate the PET by secreting the PET hydrolase (PETase) enzyme. Thus degraded PET can be used by other microorganisms as a carbon source.
Habitat of Ideonella sakaiensis
- I. sakaiensis was initially discovered from PET-contaminated soil, suggesting its main habitat to be the environment, mainly soil with enriched plastic wastes. They are found to live in oxygen-rich moist soil and sewage sludge.
Morphology of Ideonella sakaiensis
- Ideonella sakaiensis is a Gram-negative, rod-shaped, motile, non-sporing, non-pigment-producing, monotrichous bacterium. They typically measure about 1.2 to 1.5 μm in length and 0.6 to 0.8 μm in width, giving the appearance of a bacillus under the microscope.
Cultural Characteristics of Ideonella sakaiensis
- Ideonella sakaiensis is non-fastidious bacteria that can be grown in common culture mediums like nutrient agar medium and tryptic soya agar medium.
- They are aerobic, mesophilic bacteria that commonly grow at a temperature range of 15 to 42°C with optimum growth at 35±2°C (30 to 37°C) and a pH range of 5.5 to 9.0 with an optimum pH range of 7 to 7.5. (Reference: doi:10.13145/bacdive140803.20230509.8)
- In NBRC no. 802 Agar Medium, I. sakaiensis gives small, circular, raised colonies of 0.5 to 1 mm with an entire margin. The colonies are non-pigmented and creamy translucent in appearance.
- Similar colonies are found in Nutrient Medium and Trypticase soy broth agar medium. The bacterium is also grown in Tryptone soya broth.
Biochemical Characteristics of Ideonella sakaiensis
General Biochemical Test Results
| General Biochemical Characteristics | Ideonella sakaiensis |
| Catalase | Positive (+) |
| Oxidase | Positive (+) |
| Gram-staining | Gram Negative |
| Growth on 3.5% NaCl | Negative (-) |
| Indole | Negative (-) |
| Urease | Positive (+) (few show negative) |
| Nitrate reduction | Negative (-) |
| Esculin Hydrolysis | Negative (-) |
| Gelatin Hydrolysis | Positive (+) |
| Motility | Motile |
| Pigmentation | Negative (-) |
Carbohydrate/Fat/Protein/Amino Acid Utilization Tests
| Carbohydrates | Ideonella sakaiensis |
| D-Glucose Fermentation | Negative (-) |
| D-Mannitol Fermentation | Negative (-) |
| D-Mannose Assimilation | Negative (-) |
| L-Arabinose Assimilation | Negative (-) |
| N-acetyl-glucosamine Assimilation | Negative (-) |
| Maltose Assimilation | Positive (+) |
| Gloconate Assimilation | Positive (+) |
| Capric acid Assimilation | Negative (-) |
| Adipic Acid Assimilation | Negative (-) |
| Malic Acid Assimilation | Negative (-) |
| Citric Acid Assimilation | Negative (-) |
| Phenyl-acetic-acid Assimilation | Negative (-) |
| Arginine Hydrolysis | Negative (-) |
| Adipate Assimilation | Positive (+) |
Enzymatic Hydrolysis Test
| Enzymes | Ideonella sakaiensis |
| Alkaline Phosphatase | Positive (+) |
| Arginine Dihydrolase | Negative (-) |
| Alpha- and Beta-galactosidase | Negative (-) |
| Beta-glucosidase | Negative (-) |
| Alpha-glucosidase | Positive (+) |
| Gelatinase | Positive (+) |
(Reference of biochemical characteristics:
1. doi:10.13145/bacdive140803.20230509.8
2. Tanasupawat, S., Takehana, T., Yoshida, S., Hiraga, K., & Oda, K. (2016). Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). International journal of systematic and evolutionary microbiology, 66(8), 2813–2818. https://doi.org/10.1099/ijsem.0.001058)
Identification of Ideonella sakaiensis
- There is no well-devised biochemical key to identify the Ideonella sakaiensis bacterium. Phenotypic identification can be made based on the above-mentioned biochemical characteristics and microscopic observation together with the report of electron microscopy suggesting the presence of a single flagellum.
- Molecular identification using the polymerase chain reaction (PCR) is the only available accurate method to identify Ideonella sakaiensis.
Potent Application of Ideonella sakaiensis
Being a persistent and not-easily biodegradable material and due to its extensive use and discard in the environment, plastic is the most intractable waste posing a catastrophic threat to the ecosystem. Lots of strategies have been built and placed to control the bulk of plastic waste and manage the existing plastic waste; but, none of them are effective enough. In this context, a plastic-eating bacterium such as Ideonella sakaiensis can be a mighty option to aid in the process of plastic waste management.
Some potent applications of Ideonella sakaiensis can be summarized as:
- Plastic Upcycling
Upcycling is the process of converting one material into another chemically different material. I. sakaiensis can enzymatically degrade plastic, mainly PET, into other forms like mono(2-hydroxyethyl)terephthalate (MHET), ethylene glycol, etc. These products can be used by the same bacterium or other bacteria and even be modified chemically into several other products.
- Bioremediation
The biological management of environmental wastes is called bioremediation. There are scanty organisms that are found to remediate plastic waste; however, their potential to control plastic waste is questionable because of their slower rate to degrade the plastics. In this context, the newly identified bacterium, Ideonella sakaiensis can be a potential being to aid the plastic bioremediation process.
- Industrial Application
PETase and MHETase enzymes produced by Ideonella sakaiensis have potential use in industries production producing plastic products and managing solid and/or sewage wastes.
Mechanism of Plastic Degradation by Ideonella sakaiensis
- Ideonella sakaiensis is found to be able to degrade polyethylene terephthalate (PET) type plastic. This type of plastic is widely used in the manufacture of plastic bottles and packaging materials.
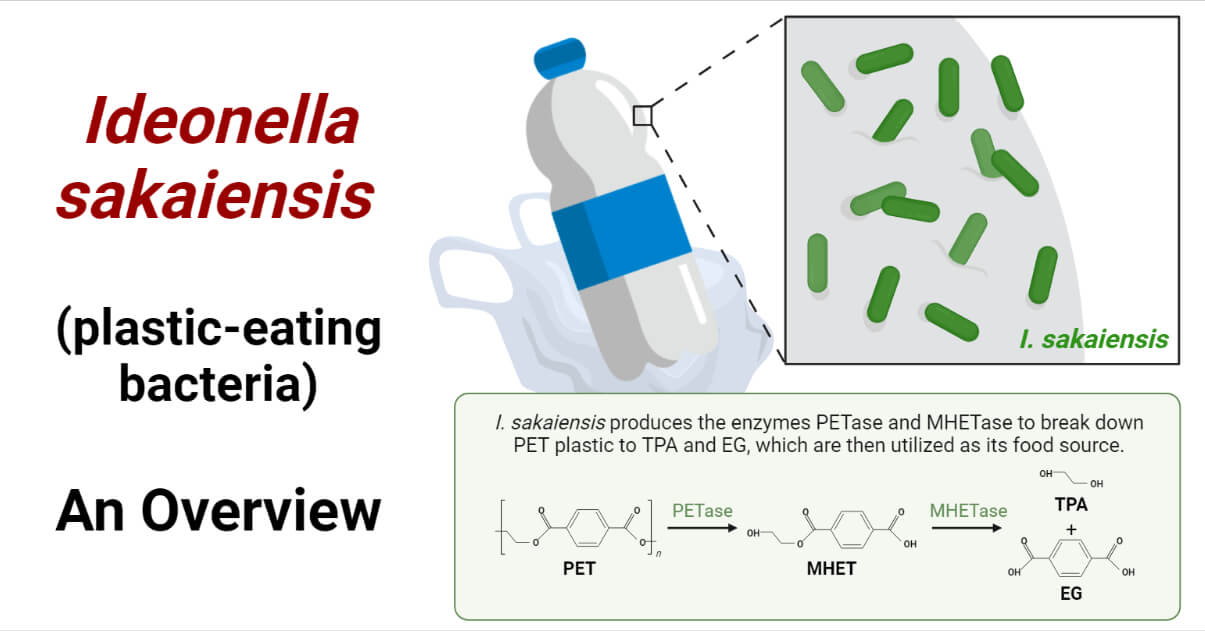
- Ideonella sakaiensis adhere to the PET plastic surface using their flagellum. Once adhered, they secret PET-degrading enzyme, the PET hydrolase (PETase) enzyme on the surface of the plastic. The PETase enzyme breaks down the PET polymer into mono (2-hydroxyethyl) terephthalic acid (MHET) and a trace amount of terephthalic acid (TPA), bis (2-hydroxyethyl) terephthalic acid (BHET), and ethylene glycol (EG) as secondary products. The BHET is an intermediate product that will be converted into MHET.
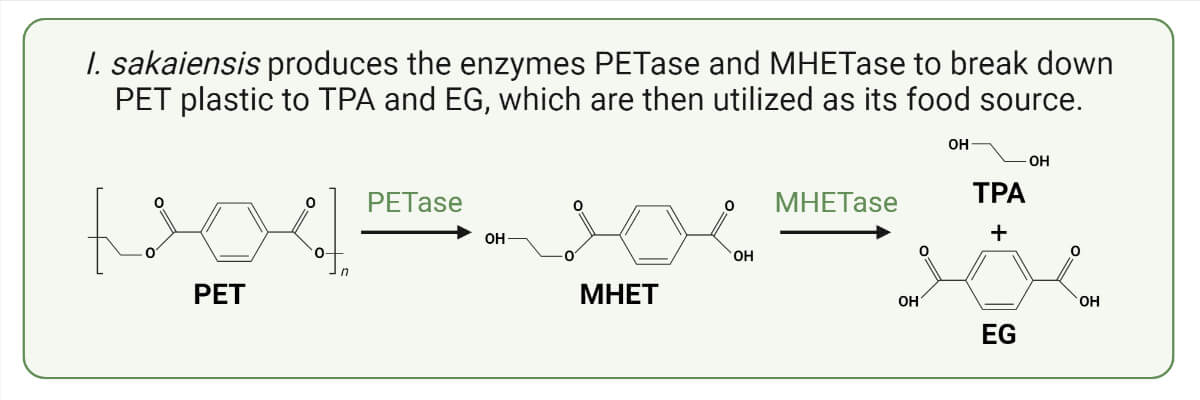
- Another enzyme secreted by Ideonella sakaiensis, the MHET-degrading enzyme commonly known as MHET hydrolase or MHETase, then converts thus formed MHET into two monomers, terephthalic acid (TPA) and ethylene glycol (EG).
- The resulting ethylene glycol is taken up by Ideonella sakaiensis and other bacteria and readily metabolized as a carbon source. Mainly they are metabolized to acetyl-CoA and then oxidized in the tricarboxylic acid (TCA) cycle to CO2 yielding reduced nicotinamides which are used in the oxidative phosphorylation process to generate the energy molecule, ATP.
The terephthalic acid (TPA) is more recalcitrant, but once inside the cell of Ideonella sakaiensis, the enzymes 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase and terephthalic acid-1,2-dioxygenase catalyze the breakdown of TPA into protocatechuate. The protocatechuate is further catalyzed and incorporated into other metabolic pathways like the TCA cycle producing ATP and CO2.
(References:
Pirillo, V., Pollegioni, L., & Molla, G. (2021). Analytical methods for the investigation of enzyme‐catalyzed degradation of polyethylene terephthalate. The Febs Journal, 288(16), 4730-4745. https://doi.org/10.1111/febs.15850
Willetts A. (1981). Bacterial metabolism of ethylene glycol. Biochimica et biophysica acta, 677(2), 194–199. https://doi.org/10.1016/0304-4165(81)90085-4
https://www.uniprot.org/uniprotkb/Q5D0X4/entry
https://www.uniprot.org/uniprotkb/Q3C1E2/entry
https://www.gbif.org/species/165596032)
There is yet to learn much about this novel bacterium’s more precise mechanism of PET hydrolysis.
References
- Tanasupawat, Somboon & Takehana, Toshihiko & Yoshida, Shosuke & Hiraga, Kazumi & Oda, Kohei. (2016). Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades PET. International journal of systematic and evolutionary microbiology. 66. 10.1099/ijsem.0.001058.
- Nishii, T., & Yoshida, S. (2021). Development of a Targeted Gene Disruption System in the Poly(Ethylene Terephthalate)-Degrading Bacterium Ideonella sakaiensis and Its Applications to PETase and MHETase Genes. Applied and Environmental Microbiology, 87(18). https://doi.org/10.1128/AEM.00020-21
- Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., Toyohara, K., Miyamoto, K., Kimura, Y., & Oda, K. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science (New York, N.Y.), 351(6278), 1196–1199. https://doi.org/10.1126/science.aad6359
- Yoshida, S., Hiraga, K., Taniguchi, I., & Oda, K. (2021). Ideonella sakaiensis, PETase, and MHETase: From identification of microbial PET degradation to enzyme characterization. Methods in enzymology, 648, 187–205. https://doi.org/10.1016/bs.mie.2020.12.007
- Willetts A. (1981). Bacterial metabolism of ethylene glycol. Biochimica et biophysica acta, 677(2), 194–199. https://doi.org/10.1016/0304-4165(81)90085-4
- Pirillo, V., Pollegioni, L., & Molla, G. (2021). Analytical methods for the investigation of enzyme‐catalyzed degradation of polyethylene terephthalate. The Febs Journal, 288(16), 4730-4745. https://doi.org/10.1111/febs.15850
- Liu, Bing; He, Lihui; Wang, Liping; Li, Tao; Li, Changcheng; Liu, Huayi; Luo, Yunzi; Bao, Rui (2018). Protein crystallography and site-direct mutagenesis analysis of the poly(ethylene terephthalate) hydrolase PETase from Ideonella sakaiensis. ChemBioChem, (), –. doi:10.1002/cbic.201800097
- Palm, G. J., Reisky, L., Böttcher, D., Müller, H., Michels, E. A., Walczak, M. C., Berndt, L., Weiss, M. S., Bornscheuer, U. T., & Weber, G. (2019). Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nature Communications, 10(1), 1-10. https://doi.org/10.1038/s41467-019-09326-3
- Widyastuti, Galuh, Genetic Engineered Ideonella Sakaiensis Bacteria: A Solution of the Legendary Plastic Waste Problem (May 27, 2018). 3rd International Conference of Integrated Intellectual Community (ICONIC) 2018, Available at SSRN: https://ssrn.com/abstract=3194556 or http://dx.doi.org/10.2139/ssrn.3194556
- I. sakaiensis – microbewiki (kenyon.edu)
- Ideonella sakaiensis. (2023, May 16). In Wikipedia. https://en.wikipedia.org/wiki/Ideonella_sakaiensis
- Ideonella – an overview | ScienceDirect Topics
- https://www.gbif.org/species/165596032
- https://lpsn.dsmz.de/genus/ideonella
- https://www.uniprot.org/uniprotkb/Q5D0X4/entry
- https://www.uniprot.org/uniprotkb/Q3C1E2/entry
- doi:10.13145/bacdive140803.20230509.8 https://bacdive.dsmz.de/strain/140803
- Ideonella sakaiensis: A Novel Method of Recycling Plastic — Teknos
