Alternative splicing is a regulated process where multiple proteins are encoded by a single gene via the joining and skipping of exons in different combinations.
In humans, alternative splicing enhances the variety of proteins encoded by the genome, this accounts for 95% of multi-exonic genes.
Interesting Science Videos
What is Alternative splicing?
Alternative splicing is a natural process where exons of a single precursor mRNA are linked in different arrangements to form two or more different variations of mature mRNAs. This strategy increases the proteome diversity and enables the control of gene expression after transcription.
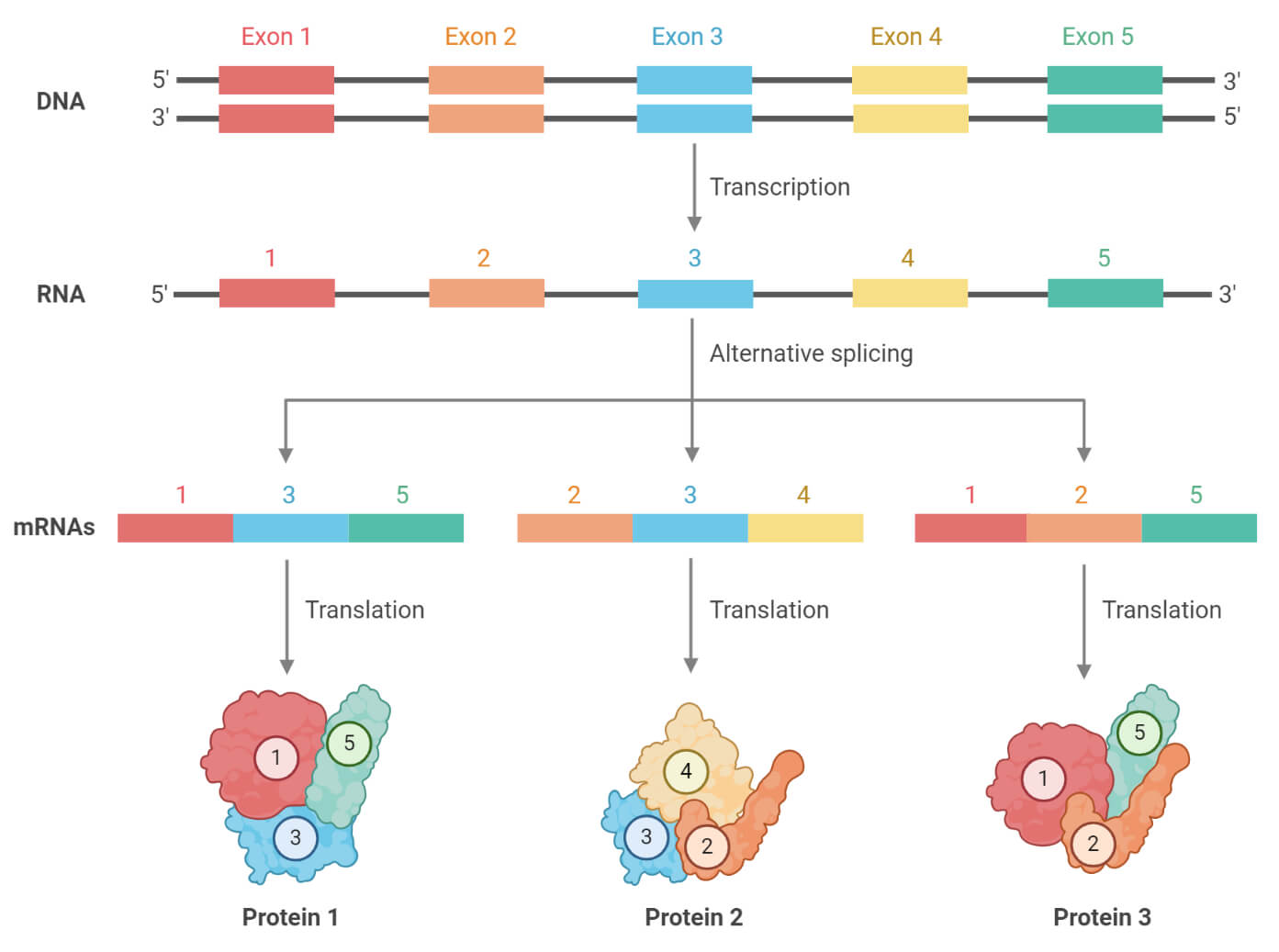
Mechanism of Alternative Splicing
- The pre-mRNA contains many introns in addition to exons.
- The exons that will be preserved in the mRNA are decided through the splicing process.
- Splice sites are regulated and selected by splicing activator and repressor proteins. Additionally, components inside the pre-mRNA, such as exonic splicing enhancers and exonic splicing silencers also regulate the splice sites.
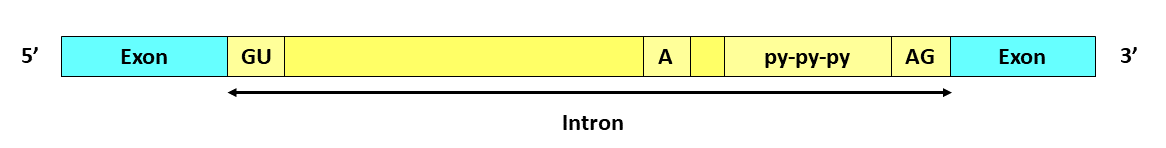
- Important areas of the nuclear intron are defined by the consensus sequences. At the 5′ end of each intron is the GU sequence.
- A branch site is located around the 3′ end. The branch site always contains adenine.
- The canonical sequence (consensus sequence) around the branchpoint differs. However, A succession of pyrimidines- polypyrimidine tract, which is further, followed by Adenine-Guanine at the 3′ end.

- A spliceosome is an RNA-protein complex that splices mRNA and contains snRNPs named U1, U2, U4, U5, and U6. (U3, on the other hand, does not involve in the mRNA splicing).
- U1 attaches to the 5′ GU sequence of the intron. Whereas U2 binds to branch point A within the branch site with the help of the U2AF protein factors. The resulting complex formed, is called spliceosome A complex.
- The forming of the A complex is one of the most important steps in selecting which intron ends should be spliced out and which exon ends should be kept.
- The U4, U5, and U6 complex bind to form tri-snRNP. U6 portrays the role of U1, whilst U1 and U4 depart the rest of the complex to perform 2 transesterification reactions mentioned below.
- Firstly, the intron is cleaved from the exon present at the 5′ end (upstream exon) and linked to branch site A by a 2′,5′-phosphodiester linkage.
- Secondly, the exon present at the 3′ end (downstream exon) of the intron is cleaved, both the cleaved exons are linked by a phosphodiester bond.
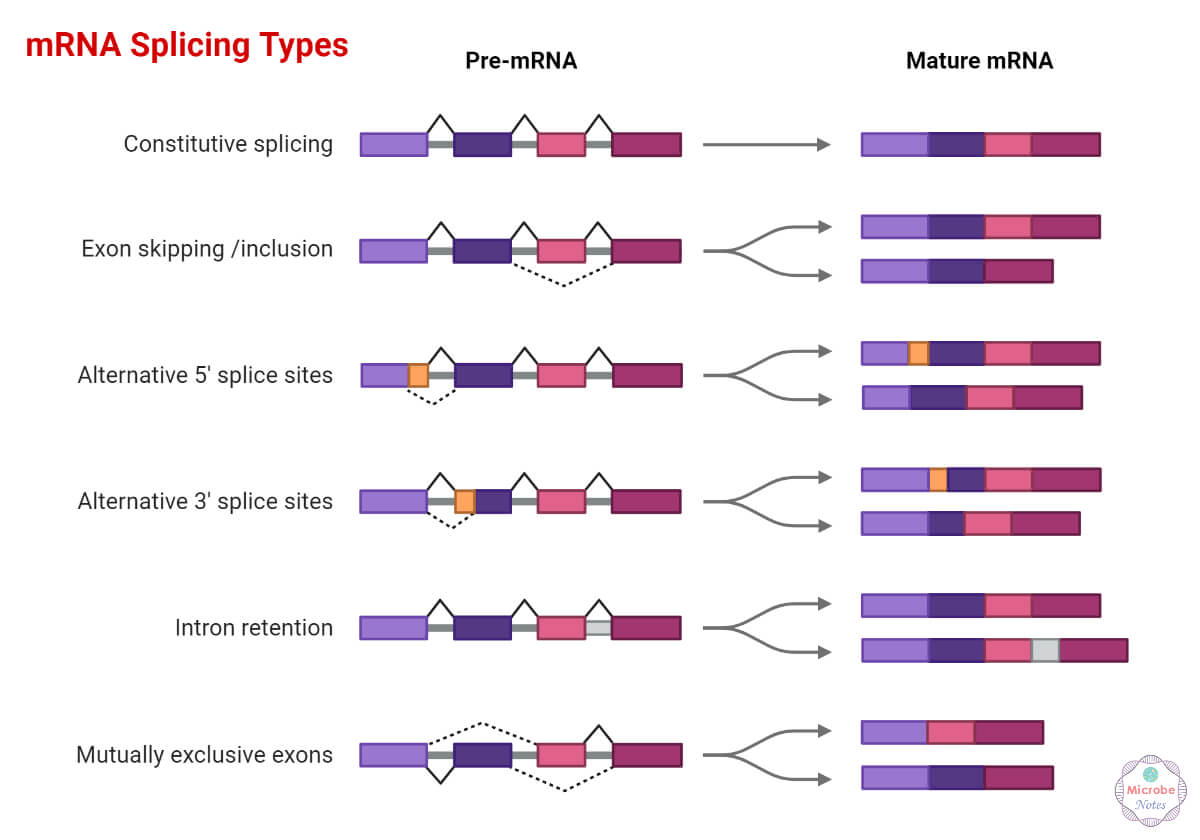
Types of Alternative Splicing
The mechanism mentioned above occurs in one of the following ways in case of alternative splicing
- Constitutive splicing: Introns are spliced, and exons are joined.
- Mutually exclusive exons: Only one of the two exons remains in mature mRNAs after splicing.
- Exon skipping or cassette exon: Exons are either spliced out of the primary transcript or maintained in mature mRNA. This is the most prevalent alternative splicing pattern (30%) in vertebrates and invertebrates.
- Alternative 3’splice site: Joining of exons at the alternate 3’splice site.
- Alternative 5’splice site: Joining of exons at the alternate 5’splice site.
- Intron retention: An intron sequence is simply retained or may be spliced out. It is the most alternative splicing prevalent pattern in lower metazoans.
Alternative splicing in Eukaryotes
A freshly transcribed RNA transcript, called pre-mRNA is not ready to be translated into eukaryotes. Instead, it has to be converted into a mature mRNA (messenger RNA) that can be translated for protein synthesis. It must go through various processing steps to mature. This includes the following
- A cap is added to the 5′ terminal of the RNA called, 5’cap.
- Addition of a poly-A tail (tail of A nucleotides) to the 3′ terminal of the RNA
- Splicing out introns, and joining together the remaining sequences (exons)
- The byproduct of these steps is a mature mRNA. The mature mRNA further moves out of the nucleus to be translated into protein formation.
Alternative splicing in Prokaryotes
In prokaryotes, mRNA does not require splicing of introns and joining of the exons, unlike that in eukaryotes.
In prokaryotes, splicing can be seen in tRNA or transfer RNA to produce its functional form with the attachment of a formalin derivative. This final step of translation attaches tRNA into mRNA along with ribosomes to form the respective protein.
Examples of Alternative Splicing
- Alternative splicing is implicated in every step of cancer development.
- Dsam gene (Drosophila Down syndrome cell adhesion molecule): it can achieve 38,016 isoforms out of 95 variable exons by alternative splicing.
- Mhc gene
- Mhc gene encodes a protein that plays a key role in the function of muscle cells
- An example of a gene that undergoes complex alternative splicing in Drosophila is the Myosin heavy chain (Mhc).
- The Mhc gene hosts 30 exons, out of that 17 are alternatively spliced, except exon 18, which is represented at the genomic level by a single alternative exon, the other alternatively spliced exons are organized into 5 separate clusters which contain about 2 to 5 exons each.
- Neurexin gene: Neurexins are presynaptic cell-adhesion molecules that are essential for synapse formation and synaptic transmission. Alternative splicing of neurexin transcript generates thousands of isoforms.
Importance of Alternative Splicing
- The main function of alternative splicing is to increase the diversify the mRNA expressed from a genome to produce a variety of proteins.
- Helps understand how numerous proteins can be generated with one gene.
- It permits exonic regulatory sequences to have a wide range of sequence freedom without compromising coding requirements.
- Slight variations in the concentration of regulatory proteins can modify protein interaction, allowing exons to be used in different ways.
References
- Han, J., Xiong, J., Wang, D. and Fu, X., 2011. Pre-mRNA splicing: where and when in the nucleus. Trends in Cell Biology, 21(6), pp.336-343. DOI: 10.1016/j.tcb.2011.03.003
- Strachan, T. and Read, A., 2001. Human molecular genetics. New York: Wiley-Liss, pp.372-373.
- Nussbaum, R., McInnes, R., Williard, H., Thompson, J. and Thompson, M., 2016. Genetics in medicine. 8th ed. Elsevier Inc, pp.32-33.
- Jung Woo Park and Brenton R; Adv Exp Med Biol. Author manuscript; PMC 2015 April 07.
- WANG, Y., LIU, J., HUANG, B., XU, Y., LI, J., HUANG, L., LIN, J., ZHANG, J., MIN, Q., YANG, W. and WANG, X., 2015. Mechanism of alternative splicing and its regulation. Biomedical Reports, 3(2), pp.152-158.