Interesting Science Videos
What is tRNA?
Definition of tRNA- Transfer RNA or tRNA is a type of RNA molecule that helps to decode information present in mRNA sequences into specific proteins.
- tRNA molecule is a carrier of amino acid that brings appropriate amino acid to ribosome based on the codon present in mRNA sequence.
- tRNA is also known as an adaptor molecule as it translates the codons present in mRNA sequences into amino acids.
- tRNA is only 70-90 nucleotides in length, making it the smallest out of the three main RNAs (mRNA and rRNA).
- The molecular weight of the tRNA molecule ranges from 25,000 to 30,000 Dalton.
- tRNA is encoded by DNA in the cell nucleus and transcribed with the help of RNA polymerase ΙΙΙ.
Properties of tRNA
- tRNA can make hydrogen bonds with mRNA and also can form ester linkage with amino acids, thus linking mRNA with the amino acid at the site of protein synthesis.
- tRNA bond complementary with each three-nucleotide of mRNA called codons and this happens in an antiparallel fashion. The part of tRNA that binds with the codon of mRNA is termed as anticodon arm of tRNA.
- This complementary base pairing of tRNA and mRNA makes the correct translation of the correct amino acid that is inserted into the growing polypeptide chain.
- The tRNA is a single-stranded RNA molecule that has the proximity to fold upon itself and creates an intra complementary base pairing which gives raise to hydrogen-bonded stems and associated loops that contains nucleotides with modified bases.
- In addition to the usual bases i.e., Adenine, Uracil, Cytosine, and Guanosine, tRNA also contains modified unusual bases like for Adenine, there is an unusual base called Inosine, likewise in place of Uracil, there are Psuedouracil and Dihydrouridine.
- These modified unusual bases and intramolecular base pairing of tRNA make it stable and protect it from getting degraded by the enzyme RNase.
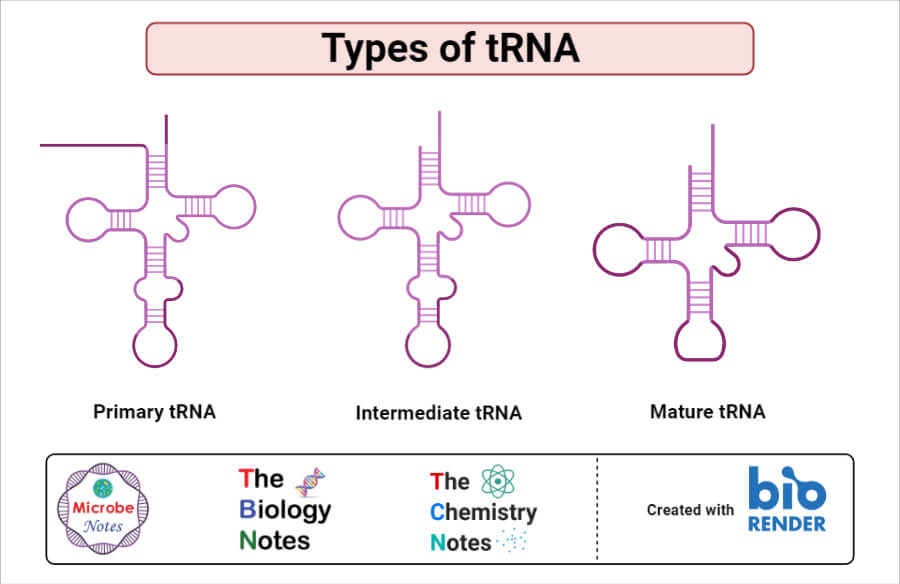
Kinds of tRNA Structure
The tRNA has three kinds of structure:
1. Primary structure
- A linear single strand of ribonucleotides running in a 5′ to 3′ direction.
- The number of ribonucleotides ranges from 60-90, the most commonly found length is 76 ribonucleotides.
- A single tRNA molecule having a primary structure consists of about 20% of modified bases.
- Based on these modified bases, the linear sequence of tRNA molecule can be divided into three arms: D arm containing a modified base Dihydrouridine, Anticodon arm containing unusual bases like Inosine derived from Adenine, or Pseudouridine from Uracil or Lysidine from Cytosine, TΨC arm containing modified base Pseudouridine.
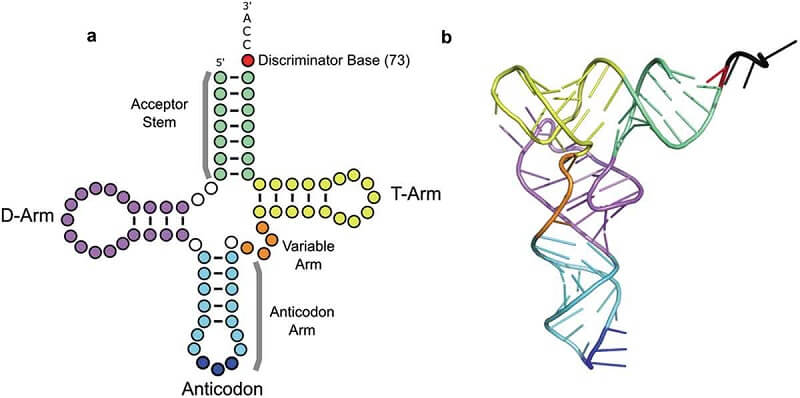
- In addition to these three arms with modified bases, an arm is present at the 3′ terminal usually containing CCA nucleotide and is referred to as Acceptor arm that acts as an attachment site for incoming amino acids.
- D arm serves as a recognition site for the enzyme aminoacyl tRNA synthetase.
- The Anticodon arm binds with the codon of mRNA during protein synthesis.
- TΨC arms contain a ribosome binding site.
2. Secondary structure (Cloverleaf model)
- The secondary structure of tRNA is of 74-95 nucleotides sequences that are stable and fold on itself to give a clover leaf-like structure containing four arms sometimes even five in longer tRNAs.
- These four base-paired arms are namely, Acceptor arm, D-arm, TΨC arm, and Anticodon arm.
- The acceptor’s arm is a 7 to 9 base pair in length, this arm is formed by base pairing of 5′ terminal and 3′ terminal nucleotides.
- Out of these four base-paired arms, three arms end up in a loop due to unpaired base-pairing forming D- loop, Anticodon loop, and TΨC loop respectively. However, the acceptor arm does not form a loop but contains an extension of the free 3′ hydroxyl group comprising of CCA nucleotides that are used to attach amino acids.
- The carboxylic group of amino acids joins with free 3′ hydroxyl of Adenine in CCA to aminoacyl tRNA, the process is also referred to as charging of tRNA and is catalyzed by an enzyme, aminoacyl tRNA synthetase.
- The acceptor arm is located opposite to Anticodon arm.
- D arm consists of a stem that is about 4 to 5 base pairs in length which end in a loop that usually contains modified pyrimidine base nucleotides, Dihydrouridine.
- D arm serves as a recognition site for the enzyme aminoacyl tRNA synthetase.
- Anticodon arm consists of a stem that is about 6 base pairs in length which end in a loop due to 7 unpaired bases. Out of which three bases form an anticodon.
- This first anticodon base is sometimes modified with unusual bases like Inosine derived from Adenine, or Pseudouridine from Uracil, or Lysidine from Cytosine.
- This anticodon base of the anticodon arm recognizes codon in mRNA and binds to it during protein synthesis.
- The TΨC arm consists of a stem that is 5 base pairs long which ends in a loop that further contains 7 unpaired base pairs, usually containing Thymine, Pseudouridine and Cytosine hence the name TΨC arm.
- The TΨC loop contains a ribosome binding site.
- In addition to four base-paired arms, a variable arm can usually be present in long tRNAs. These variable arms vary in length from 3-21 nucleotides depending on the type of amino acid they code.
- The variable arm lies between the anticodon arm and TΨC arm, and is usually short and looks sort of different from the regular four arms. The base pairing is rarely seen in this arm, so it appears as loops due to unpaired bases.
- The variable arm accounts for the stability of a tRNA molecule.
3. Tertiary structure
- It is a three-dimensional structure, where tRNA adopts an L-shaped structure.
- This L-shape is formed when the stem of acceptor and TΨC arms stack at one side and stem of anticodon and D arms stack on another side and both forms extended helices.
- Both of these extended helices align at a right angle, and in doing so, the D-loop and TΨC loop align together.
- The tertiary structure is the most stable structure out of all, and this stability is acquired due to hydrogen bonding in between nitrogenous bases and also between nitrogenous bases and the ribose-phosphate backbone.
- To synthesize proteins by reading codon in mRNA a tRNA molecule must have a tertiary structure.
Composition of tRNA
- tRNA being a type of RNA molecule is composed of a ribose sugar, phosphate, and nitrogenous base.
- It is a biopolymer of ribonucleotides.
- tRNAs are transcribed from the genes present in DNA. And as we know, DNA runs from a 5′ end to a 3′ end, so tRNA that is synthesized does have the same direction.
- During the processing of the precursor tRNA to mature tRNA, many additions and modifications are made, hence
- These modified bases in a tRNA structure characterize distinct portions in the chains of ribonucleotides.
- Based on these modified bases, tRNA molecule has three arms or call it segments, one being a D arm that distinctively has an unusual base called Dihydrouridine derived from the methylation of Uracil, another one being TΨC arm that is characterized by the presence of another methylated Uracil called Pseudouridine and the final one is Anticodon arm that has a modified base of Adenine, called Inosine.
- Along with these three distinct arms, an important arm is also present that does not contain modified bases but after processing becomes an acceptor arm that attaches amino acids.
- In summary, a tRNA molecule is composed of chains of ribonucleotides, in which certain modification of bases and addition of nucleotides takes place during processing resulting in the formation of four arms: D arm, TΨC arm, Anticodon arm, and Acceptor arm.
Processing of tRNA
- Like all RNA, tRNA is synthesized by transcribing the genes encoded in the DNA molecule with the help of an enzyme called RNA polymerase ΙΙΙ.
- And are made translation-ready (matured) through splicing, CCA addition, and methylation of bases in nucleotides.
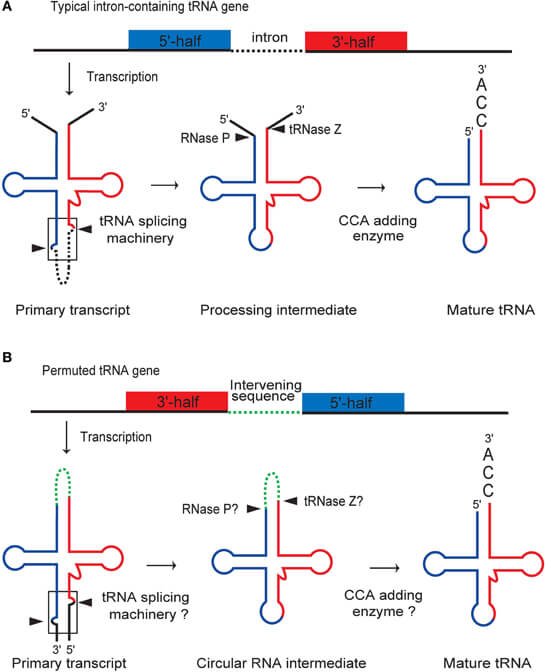
- The newly transcribed precursor tRNA has extra sequences at the 5′ and 3′ ends that are cleaved off by the action of an enzyme called RNase P, which is a ribonuclease.
- In some cases, precursor tRNA also contains a non-coding region called introns, which are spliced out by a specific endonuclease, and then the remaining fragments are joined with help of RNA ligase.
- CCA addition at the 3′ end is done with the help of an enzyme called tRNA nucleotidyltransferase.
- Modification of bases also takes place during the processing of precursor tRNA and is made possible by an enzyme, tRNA methylase that helps to provide a methyl donor, thus making the bases methylated which gives rise to modified unusual bases like Pseudouridine, Inosine, Dihydrouridine, and Lysidine.
- Each modification is made to functionalize tRNA for protein synthesis.
- Addition of CCA nucleotides at 3′ end help to attach specific amino acid by forming ester bond (esterification) with a free hydroxyl group of Adenine nucleotide of CCA. This reaction is catalyzed by an enzyme aminoacyl tRNA synthetase.
- Splicing makes the tRNA free from introns. And methylation of bases makes them recognizable by aminoacyl tRNA synthetase and ribosome.
- Hence, all this processing must take place, to make mature tRNA that can further be beneficial during protein synthesis.
Types of tRNA
- The types of tRNA are categorized on the types of amino acids it carries. A human typically has 20 essential amino acids which thus give rise to 20 different types of tRNA.
- However, alternatively, they can also be categorized based on their anticodons.
- When genes are expressed to proteins. DNA is transcribed to mRNA which is then translated into protein by tRNA. DNA contains four nucleotides so their possible combination gives rise to 64 codons.
- Out of 64 codons, 3 codons are stop codons i.e., UAG, UAA, and UGA. These three codons trigger the termination of translation. In addition to tRNA molecules being needed to pair with each amino acid. Some other tRNA molecules are also needed to pair up with these stop codons to halt the protein synthesis.
- Some codons code for the same amino acid, thus the real number of tRNA is not 64. This redundancy in genetic code is referred to as “wobble”.
- Due to this redundancy, very few species of organisms have exactly 61 codons that code for amino acids.
- tRNA interacts with codons via its anticodon loop. Base pairing between anticodon and codon ensures the specificity of amino acids to be incorporated in polypeptide chains.
Functions of tRNA
- tRNAs are vital molecules taking part in protein synthesis that transport amino acids to the ribosome where these amino acids are added according to the codon sequences encoded in the mRNA molecule.
- Picks up specific amino acids in the cytoplasm and carries them to the site of protein synthesis.
- Also participate in non-protein synthetic processes, such as primer during the reverse transcription of retroviruses.
- It is also known as an adaptor molecule because it attaches specific amino acids that the codon represents in the mRNA sequence. It can also be referred to as a bridge that links amino acids and nucleotides.
- Mutations in tRNA or tRNA processing are linked with various human diseases, like cancer and neurodegenerative diseases.
- In recent times, the tRNA molecules are altered to accommodate desired anticodons that result in translating unusual amino acids that do some specific functions. For example, the tRNA-dependent aminoacylation of bacterial cell membrane lipids has accounted for increased virulence of bacteria and their resistance against antibiotics.
tRNA Frequently Asked Questions (FAQs)
Q. What is tRNA?
Ans. Transfer RNA or tRNA is a type of RNA molecule that helps to decode information present in mRNA sequences into specific proteins.
Q. What is the composition of tRNA?
Ans. tRNA is composed of a ribose sugar, phosphate, and nitrogenous base.
Q. What are the 3 kinds of tRNA structure?
Ans. tRNA can be divided into 3 kinds of structures- Primary structure, Secondary structure (Cloverleaf model), and Tertiary structure.
Q. How many tRNAs are there in humans?
Ans. There are 22 different tRNAs in human mitochondria.
Q. Where is the tRNA produced?
Ans. In Eukaryotes, Mature tRNA is produced (generated) in the Nucleus.
Q. What are the functions of tRNA?
Ans. tRNA helps in protein synthesis that transports amino acids to the ribosome where these amino acids are added according to the codon sequences encoded in the mRNA molecule.
References
- Soma A. Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development. Front Genet. 2014 Apr 1;5:63. doi: 10.3389/fgene.2014.00063. PMID: 24744771; PMCID: PMC3978253.
- Hori, H., Tomikawa, C., Hirata, A., Toh, Y., Tomita, K., Ueda, T. and Watanabe, K. (2014). Transfer RNA Synthesis and Regulation. In eLS, John Wiley & Sons, Ltd (Ed.).
- https://www.britannica.com/science/transfer-RNA
- https://biologydictionary.net/trna/
- https://www.nature.com/scitable/definition/trna-transfer-rna-256/
- https://courses.lumenlearning.com/microbiology/chapter/structure-and-function-of-rna/
- https://www.slideshare.net/manjeshsaakre/trna-structure-and-function
- http://dnaofbioscience.blogspot.com/2016/05/clover-leaf-model-of-t-rna.html
- https://en.wikibooks.org/wiki/Structural_Biochemistry/Nucleic_Acid/RNA/Transfer_RNA_(tRNA)
- https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Boundless)/7%3A_Microbial_Genetics/7.06%3A_Translation%3A_Protein_Synthesis/7.6A%3A_Processing_of_tRNAs_and_rRNAs
- https://courses.lumenlearning.com/wm-biology1/chapter/reading-trna/
- O’Donoghue, P., Ling, J., & Söll, D. (2018). Transfer RNA function and evolution. RNA biology, 15(4-5), 423–426. https://doi.org/10.1080/15476286.2018.1478942
- Kirchner, S., Ignatova, Z. Emerging roles of tRNA in adaptive translation, signaling dynamics, and disease. Nat Rev Genet 16, 98–112 (2015). https://doi.org/10.1038/nrg3861

The author explained very well on the tRNA. helpful
thank you for this note, it really helps