The term synapomorphy is derived from the ancient Greek terms, ‘syn’ meaning ‘with, together’, ‘apo’ which means ‘moving away’, and ‘morphe’ which refers to ‘form or shape’. It was coined by Will Hennig, a German entomologist. In phylogenetic studies, when an advanced trait or character is shared among two or more closely related taxa implying their inheritance from the most recent common ancestor that the individual taxon evolved from is considered as Synapomorphy. The ancestor is inferred not to have such an advanced trait and the groups derived it through evolution.
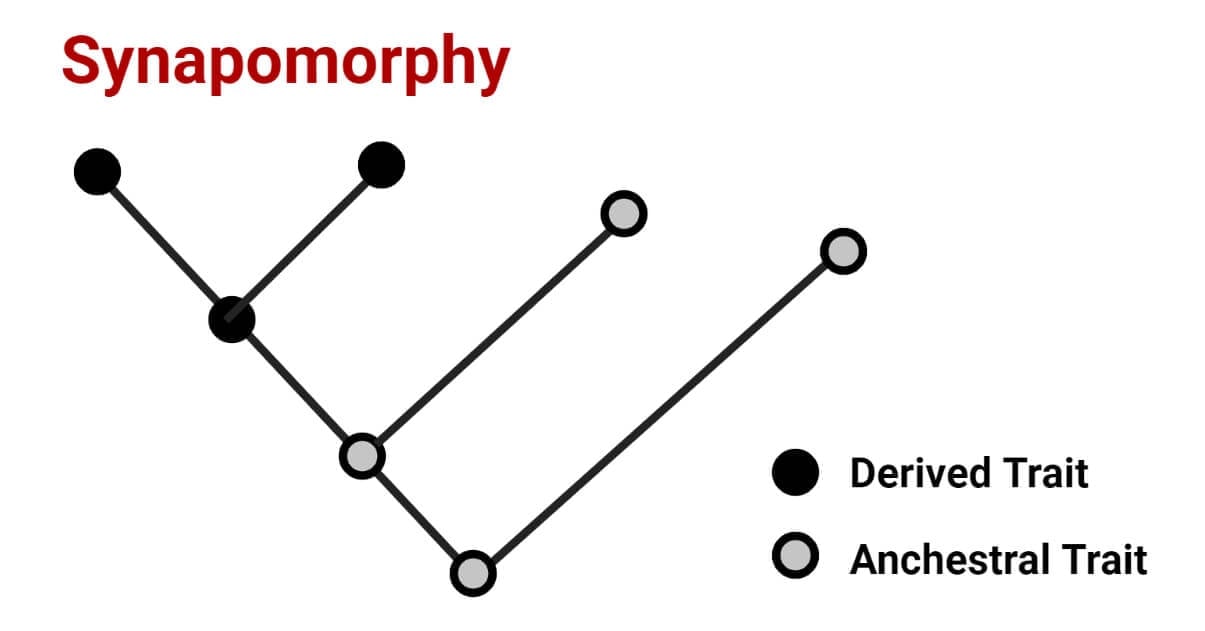
Synapomorphy combines a set of groups that own a similar character with a modified version but do not include all descendants of a clade. The derived modified trait in a group of organisms is anything observable like the type of scale or skin covering an organism possess, the colour or shape of an eye, or the size of an organism in that particular group. In modern phylogenetic studies, a specific DNA sequence approach is also considered a trait for synapomorphy classification. This helps the scientist to classify organisms as being related or not.
Importance of Synapomorphy
Synapomorphy greatly signifies how two or more organisms are closely related to each other in the phylogenetic profile. With the discovery of more ancient organisms and their fossils that roamed Earth’s surface at different eras, there is a need for evolutionary trees and cladogram formation to observe any connecting link between the discovered fossil and the modern existing species. The modern animal and plant species we see today on our planet are different from their ancestors and were not always present since the beginning of time. Synapomorphy help study the evolutionary event that led to the adaptation of a new modified trait in a group from the old similar trait seen in the recent ancestral organism.
Cladistic Analysis
- In Cladistics, synapomorphy is related to homology. The synapomorphy concept is applied to a clade in forming the tree of life, Cladograms.
- Cladograms depict evolutionary relationships among taxonomical groups, which shows how species are linked to one another.
- Synapomorphy creates evidence for ancestral relationship and their hierarchical status.
- The synapomorphic trait is set as a marker for the recent most common ancestor of a particular monophyletic group with similar advanced characters.
- For instance, the presence of a mammary gland is a synapomorphic trait exclusively for mammals.
Synapomorphy Examples
- The category of ‘apes’ include gorillas, chimpanzees, humans, and orangutans. This synapomorphic category is distinct from other primates due to the presence of traits like grinding teeth, flatter rib cages, mobile shoulder joints used for swinging from the branches, and the lack of a tail. Other mammals and primates like monkeys are devoid of these features, which depicts recent common ancestral evolutionary links for apes.
- The appearance of forearm bones in cats and humans (carnivores and primates respectively) are very similar in morphological structure. This indicates that even though they evolved via different evolutionary paths, they shared a common ancestor in the past which led to acquiring these common traits.
- Synapomorphy trait of the vertebrate category is the existence of vertebrate column that helps in supporting limbs and evenly distributing the weight of an organism, suggesting the animals are linked to one another and had a common ancestor.
- The synapomorphic trait of vascular plants but seedless which includes club mosses, horsetails, whisk ferns, etc, consists of tissues specialized for the vascular conduction of materials. Tracheids are the main conducting system of water in the xylem.
- The synapomorphy trait of multicellular sporophyte i.e. the spore-producing stage is observed in the land plants category that includes conifers, angiosperms (flowering plants), and liverworts (flowerless plants) but excludes Coleochaete which is a type of algae.
Apomorphy vs. Synapomorphy
- The term ‘apomorphy’ is used to describe any shared trait or character visible between two or more taxonomic groups of organisms.
- And the conversion of apomorphy to the synapomorphic category is possible when the shared trait is inherited from a recent common ancestor.
- The ancestral evidence must be present in the fossil record for studying and analysing the evolutionary patterns opted for producing the modern organism we see today.
- Synapomorphy signifies the relatedness among species with similar traits with a single common ancestor.
- Two or more organisms showing synapomorphic traits lead to a formation of a clade which is used to describe phylogenetic relationships among organisms.
Plesiomorphy vs. Synapomorphy
Plesiomorphy term is used when the shared character among two organisms is inherited from different ancestors as opposed to synapomorphy where the groups of organisms and the common recent ancestor, all share a similar trait.
Homoplasy vs. Synapomorphy
- The term homoplasy in phylogenetic studies is the opposite of synapomorphy or homology.
- A synapomorphic trait signifies a homologous character inherited from a recent common ancestor that is present in all organisms of a clade.
- Whereas homoplasy is referred to when a similar character is applied to different organisms of different clades.
- This is very common in evolutionary history as different species evolve to adapt to the same task due to factors like environmental conditions.
- For example, an adaptation of wings by different organisms like birds and insects cannot be considered a synapomorphy but is homoplasy because the trait is similar from different ancestors acquired in the course of evolution.
- Similarly, the wings of bats and birds are a homoplasy trait.
Conclusion
Synapomorphy is a useful tool for understanding the evolutionary patterns and relationships among different groups of organisms and helps reconstruct the tree of life. The term is used in evolutionary biology referring to a shared trait or character inherited from a recent common ancestor which is unique to a particular clade of organisms. To be included in the synapomorphy category, a character must meet the criteria of being present in all organisms of that particular clade but absent in organisms outside of a clade and must be derived from a common ancestor that evolves in the lineage to the clade/group of interest. Phylogenetic trees are constructed based on the synapomorphic traits providing strong evidence for shared ancestry among organisms. Synapomorphy infers relatedness among organisms that evolved over time. Some examples include the presence of feathers in birds, hair in mammals, etc.
References
- Apomorphy and synapomorphy – https://en.wikipedia.org/wiki/Apomorphy_and_synapomorphy
- Synapomorphy: Understanding the Definition Through Apt Examples – https://biologywise.com/synapomorphy-definition-examples
- Synapomorphy – https://biologydictionary.net/synapomorphy/
- Synapomorphy – https://www.biologyonline.com/dictionary/synapomorphy
- Plant Diversity – http://www.eebweb.arizona.edu/courses/182-spring2009-bonine/182-keb-plantdiversity-lect4-sp2009postx6.pdf
