Polymerase Chain Reaction (PCR) is a nucleic acid amplification technique used to amplify the DNA or RNA in vitro enzymatically. It is a temperature-dependent enzymatic process where either a specific targeted region of DNA or the whole DNA is replicated to quickly make millions of copies of the target DNA or DNA segment. It is also called “molecular photocopying”.
It is a non-cultured based nucleic acid cloning technique. We can quickly produce billions of copies of a single segment of DNA or RNA using the PCR technique. It is temperature dependent cyclic process, and by the end of the 20th to 25th cycle, over a million copies of the target nucleic acid segment will be produced.
It combines the principle of nucleic acid hybridization with the principle of nucleic acid replication. It denatures the double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA), selects a segment of DNA to amplify using a specific primer, and then follows the method of replication using DNA polymerase enzyme to amplify that segment.
PCR dates back to the mid-1980s when Kary Mullis and his associates developed this revolutionary technique. He was awarded the Nobel Prize in chemistry with Michael Smith in 1993. Since then, it has been the most important tool in molecular biology and genetics as a basic tool for DNA and RNA analysis.
Objectives of Polymerase Chain Reaction (PCR)
- To amplify the specific segment of DNA or RNA, resulting in billions of copies of a single DNA or RNA segment.
- To reduce the time required for the cloning of nucleic acids
- Additionally, it has been modified to diagnose certain genes, and mutations, and support other genetic and molecular studies or techniques
Principle of PCR
PCR combines the principle of nucleic acid hybridization with the principle of nucleic acid replication with the temperature variations of cyclic heating and cooling throughout the process. It is based on the fact that double-stranded DNA can be thermally broken into single-stranded DNA segments. In these ssDNA templates, primers can anneal to their complementary sequences based on the nucleic acid hybridization principle. DNA polymerase then elongates the primer by sequentially adding the nucleotides to the 3’ end and generates a dsDNA following the principle of DNA replication. By continuously regulating the temperature, these 3 steps; denaturation, annealing, and elongation, can be permitted to occur in a cyclic manner leading to millions of copies of a single targeted nucleic acid sequence.
Components of PCR (Enzymes used in PCR)
1. Nucleic Acid Template (Template DNA)
Template DNA is the sample DNA that contains the selected nucleic acid sequence that needs to be amplified. The template must be DNA only: Genomic DNA (gDNA), complementary DNA (cDNA), and plasmid DNA. Reverse transcriptase polymerase chain reaction (RT-PCR) uses RNAs as starting materials, but RNAs are primarily converted to complementary DNA (cDNA) before amplification. The template DNA must be highly pure with an absorbance ratio of ~1.8. A quantity of 0.1 to 200 μg can be used, with an ideal quantity of 30 μg to 50 μg.
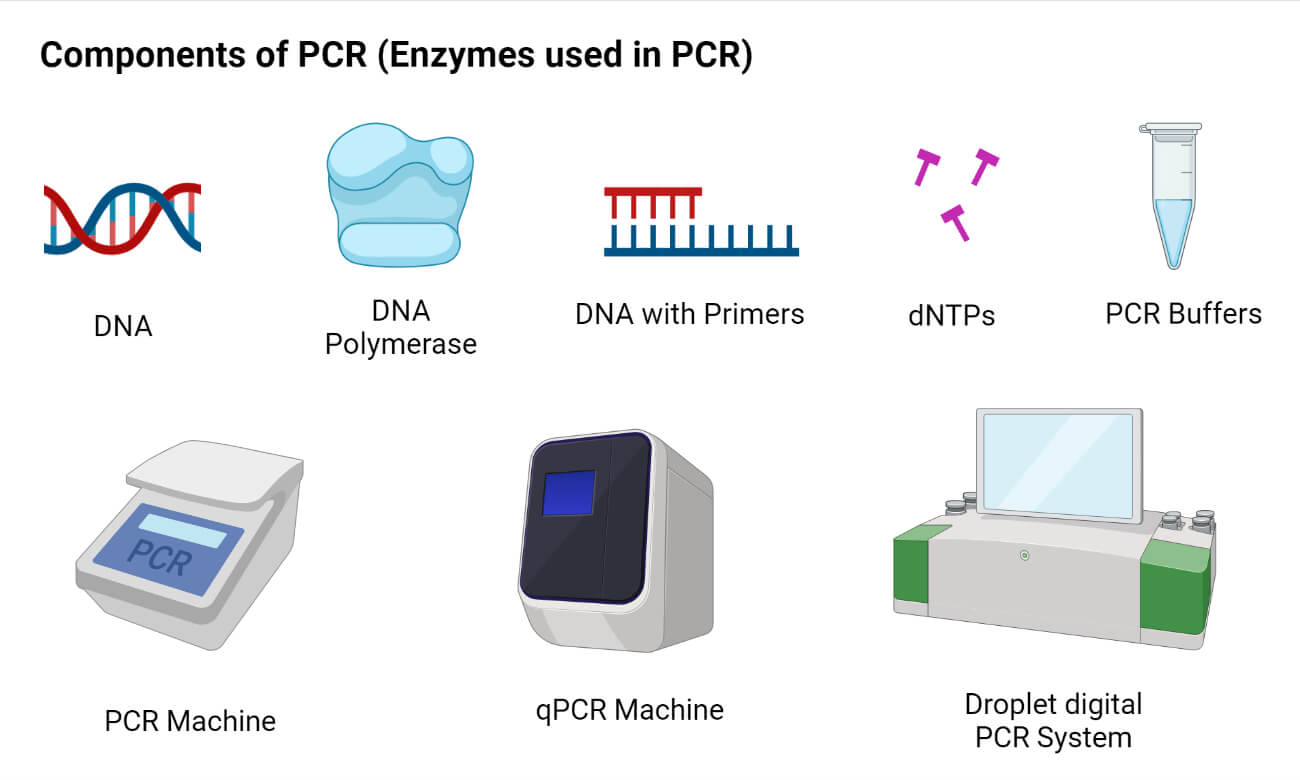
2. DNA Polymerase
DNA polymerases are enzymes that catalyze the synthesis of complementary DNA strands by assembling the nucleotides sequentially according to the template strand. Simply, it is the enzyme that synthesizes DNA; hence plays a key role in DNA replication.
Taq DNA polymerase, the DNA polymerase enzyme extracted from the bacterium Thermus aquaticus, is the most widely and the best–known DNA polymerase used in PCR since its establishment. Taq DNA polymerase is thermally stable and continues its activity after the repeated heating and cooling cycle. It is stable up to 95°C and shows the most effective reaction at around 72°C to 78°C incorporating about 60 bases per second. In a 50 L reaction mixture, around 1 to 2 units of Taq polymerase is sufficient for amplification.
Recently, two other thermostable DNA polymerase enzymes are available, viz., the Vent enzyme isolated from Thermococcus litoralis, and the Pfu enzyme isolated from Pyrococcus furiosus.
3. Primers
Primers are artificially synthesized short single-stranded sequences of oligonucleotides that are complementary to the target nucleic acid sequence in the template DNA. They are short sequences of around 15 to 30 bases that act as starting point for DNA synthesis. They anneal at their complementary position in a single-stranded template DNA strand. The DNA polymerase enzyme then extends this primer from its 3’ OH- end forming a new complementary strand. Usually, 10 to 12 pMol of each primer is sufficient for a PCR reaction.
PCR primers are of 2 types; forward and reverse primers. The forward primers are complementary to the antisense strand (template strand from 3’ to 5’ direction), and are responsible for the amplification of the antisense strand. They are also called 5’ primers.
The reverse primers are complementary to the sense strand (template strand from 5’ to 3’ direction) and are responsible for the amplification of the sense strand. They are also called 3’ primers.
4. Nucleotides (Deoxynucleotide triphosphates)
Deoxynucleotide triphosphates (dNTPs) are artificially synthesized nucleotides that act as building blocks of new DNA strands. There are 4 different dNTPs used in the PCR; deoxyadenosine triphosphate (dATP), deoxyguanosine triphosphate (dGTP), deoxythymidine triphosphate (dTTP), and Deoxycytidine triphosphate (dCTP). These four dNTPs are sequentially added to the annealed primer by the DNA polymerase enzyme generating a new strand of DNA complementary to the template strand.
5. PCR Buffers and Other Chemicals
The whole process needs to be carried out in a Tris – HCl based buffer system of pH 8.0 to 9.5. The common buffer system used is a 10X buffer with additional MgCl2. Common components of PCR buffers are dimethyl sulfoxide (DMSO), ammonium sulfate ((NH4)2SO4), nonionic detergents, polyethylene glycon(PEG), N,N,N-trimethyl glycine, potassium chloride (KCl), magnesium chloride (MgCl2), tetra methyl ammonium chloride, Tris – HCl, Ethylenediaminetetraacetic acid (EDTA), 7-deaza-2′-deoxyguanosine 5’-triphosphate, glycerol, formamide, serum albumin, etc. The buffer system increases the reaction’s efficiency and specificity and prevents inhibition and secondary structure formation during the process.
6. Thermocycler
Also known as the PCR machine, the thermocycler is simply an electric heating device that regulates the temperature as per need during each stage of the PCR process. This machine increases the temperature during the denaturation step and lowers it during the annealing and again increases it during the elongation step. This process of increasing and decreasing the temperature occurs in a cyclic manner according to the pre-programmed setup or instruction by the user prior to operating.
Steps of PCR
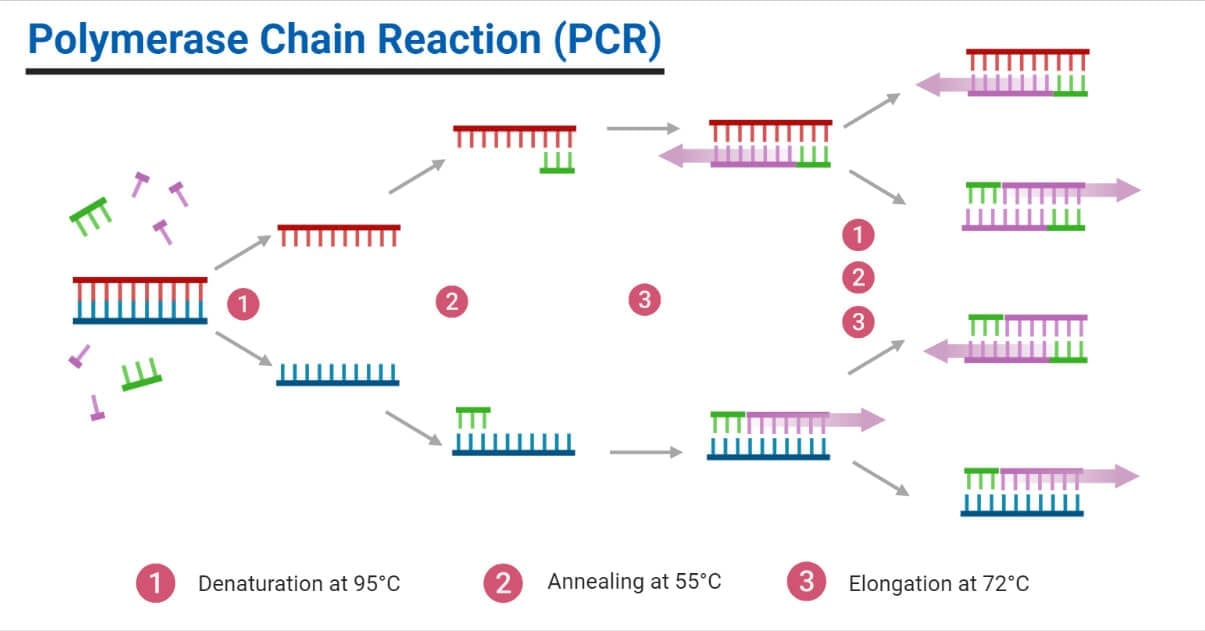
There are three core steps in any type of PCR: denaturation, annealing, and elongation. Apart from these three primary steps, the preparation phase at the beginning and the PCR product analysis phase at the end are also important steps. The whole steps can be summarized into three steps; pre-preparation, amplification, and product analysis.
1. Pre-preparation
It is the initial step before the actual polymerase chain reaction takes place inside the thermocycler. In this step, one must prepare a reaction mixture and load it on a pre-programmed thermocycler in order to amplify the target DNA or RNA segment. Sample DNAs or RNAs are extracted from the sample and stored (pre-extracted nucleic acids can be used). All materials are arranged, safety measures are taken, the PCR reaction preparation area is cleaned, all the reagents are brought to working temperature, the sample is extracted or brought from storage, the PCR reaction mixture is prepared, the thermocycler is programmed, and the reaction mixture is loaded on the thermocycler.
2. Amplification
It is the main reaction process occurring in PCR. The amplification step includes denaturation, annealing, and elongation occurring orderly in a cyclic manner one after another for a certain number of cycles pre-programmed by the user.
Step I: Denaturation
It is the 1st step of the amplification reaction where the double-stranded DNA is thermally denatured into two single-stranded DNA templates. Temperature is raised to about 94°C (90 to 95°C) for about 30 to 90 seconds. At this temperature, the thermal energy overcomes the weak hydrogen bonds holding the two DNA strands together, allowing them to separate.
dsDNA → 2 ssDNA templates
Step II: Annealing
Denaturation is followed by the annealing step, where the primer anneals the ssDNA templates at their complementary sites. The forward primer anneals at the complementary site of the antisense strand, and the reverse primer anneals at the complementary site of the sense strand of the template DNA. For annealing to occur, the temperature is reduced to 55°C-70°C (the annealing temperature differs based on the GC content of the primer). About 30 to 60 seconds are enough for annealing in most of the PCR processes.
ssDNA + Forward and reverse primers → ssDNA with annealed primers
Step III: Elongation
It is the final step in the amplification reaction where the temperature is raised to 72°C so that the Taq DNA polymerase enzyme begins synthesizing new DNA strands in the 5’ to 3’ direction. The DNA polymerase enzyme adds nucleotides from the reaction mixture to the 3’ OH- end of the annealed primer forming a new complementary strand. The time required for elongation depends on the sample nucleic acid sequence length and the DNA polymerase activity. Generally, elongation takes place at the rate of 1 kbp per 0.5 to 1 minute. At the end of elongation, two new dsDNA will be formed from a single dsDNA template at the beginning of the reaction.
2 ssDNA with annealed primers + dNTPs → 2 new dsDNAs
3. Product Analysis Phase
It is the phase after completion of the PCR where the reaction mixture subjected to PCR is analyzed to confirm that desired amplification is achieved. For this, mostly agarose gel electrophoresis is employed in order to check for amplified DNAs or RNAs. However, no additional step is required in some types of PCR, like real-time PCR.
Types of PCR
- Arbitrarily Primed PCR
- Allelic specific PCR
- Asymmetric PCR
- Assemble PCR
- Amplified Fragment Length Polymorphism PCR
- Alu PCR
- Conventional PCR
- COLD PCR
- Colony PCR
- Digital PCR
- Droplet PCR
- Fast Cycling PCR
- Gradient PCR
- GC- Rich PCR
- High Fidelity PCR
- Hot Start PCR
- High-Resolution Melt PCR
- Intra-sequence Specific PCR
- In Situ PCR
- Immuno PCR
- In Silico PCR
- Inverse PCR
- Long Range PCR
- Ligation Mediated PCR
- Linear After the Exponential PCR
- Multiplex PCR Real-time PCR
- Methylation Specific PCR
- Mini prime PCR
- Nanoparticle Assisted PCR
- Nested PCR
- Overlap Extension PCR
- Quantitative Real-Time PCR
- Repetitive Sequence-Based PCR
- Reverse Transcriptase PCR
- Real-Time PCR
- Reverse Transcriptase Real-Time PCR
- RNase H Dependent PCR
- Single Cell PCR
- Single Specific Primer PCR
- Solid Phase PCR
- Suicide PCR
- Thermal Asymmetric Interlaced PCR
- Touch Down PCR
- Variable Number of Tandem Repeats PCR
Polymerase Chain Reaction (PCR) Applications
Since its establishment, PCR has found numerous applications in different aspects of molecular biology. Some of them are;
1. Identification and Classification of Organism
PCR is used widely in identifying microorganisms up to the level of subspecies and strains. This has reduced the time required for microbial identification from days to a few hours. Additionally, larger animals can also be identified and systematically classified using PCR. DNA isolated from fossilized animals are also amplified and studied to relate them with animals that are still living on the Earth.
2. Infectious Disease Diagnosis
The use of PCR in the identification of pathogens has led to the quick and accurate diagnosis of infections. Not only diagnosis but parallel identification of antimicrobial resistant genes in the pathogen is also possible, allowing choosing of appropriate antimicrobial treatment option.
HIV, SARS CoV – 2, human T – cell leukemia virus (HTLV type I and II), Tuberculosis, Hepatitis Virus, Enterovirus, Sexually Transmitted Diseases (STDs), etc., are diagnosed using PCR.
3. Detection of Gene Mutation and Genetic Disorders
Mutation in any segment of a gene can be detected using PCR. Knowing this mutation, we can confirm a genetic disorder. DNA polymorphism can also be analyzed, which can also relate to a genetic disorder of some kind. In medical science, PCR is used as one of the most important tools to diagnose congenital diseases, genetic disorders, and any mutation leading to a negative health problem and behavioral change in the prenatal stage. Detection of cancerous cells is another very important application of PCR in medicine.
4. DNA Fingerprinting
In forensics, PCR is used for DNA fingerprinting. DNA fingerprinting is used for the identification of criminals or individuals and for confirming parents.
5. Gene Sequencing
For gene sequencing, a gene must be amplified into a large number using techniques like PCR. All the sequencing methods use PCR as their important step.
6. DNA and RNA Quantification
PCR can also be used for the quantification of sample DNA and RNA. Quantitative Real-Time PCR (RT – qPCR) is one common type of PCR used for the quantification of sample DNA.
7. As a Tool in Genetic Engineering
PCR is used in genetic engineering for analyzing modified DNAs and amplifying target or vector DNA. Desired genes are amplified using PCR and applied in the required process.
8. Gene Expression Analysis and Genetic Imprinting
PCR of RNA (Reverse Transcription PCR) is used in gene expression analysis, study genetic imprinting, etc.
9. It is also used in drug and vaccine discovery, human genome projects, paleontology, and evolutionary biology.
Advantages of Polymerase Chain Reaction (PCR)
- It is a very fast method for producing multiple copies of a nucleic acid segment. Within a few hours to a few days, billions of copies of a specific nucleic acid sequence can be produced and analyzed.
- It can analyze a very minute quantity, 0.1 to 5 μg, of DNA or RNA.
- Its principle and procedure are simple to understand and apply. Being a semiautomatic technique, the process is operated and regulated automatically by a thermocycler.
- It has shortened the time for the diagnosis of diseases and genetic disorders.
- It is the base for several molecular techniques like DNA fingerprinting, genetic imprinting, microarray, gene sequencing, molecular vector production, etc.
Limitations of PCR
- Prior information regarding the sequence of the DNA or RNA target segment is required. Only then we can design and generate primers and selectively amplify the target.
- Even very small contamination in a sample can be amplified, giving a false positive or false negative result.
- There is a chance of mutation during the process if the DNA polymerase enzyme makes any error during replication.
- PCR fails to detect novel mutations, multigenic disorders, and structural and numerical chromosomal abnormalities.
- Several organic or inorganic contaminants can disrupt the process, inhibiting the process and producing inaccurate results.
- It is a highly temperature-dependent process; hence there must be strict regulation of temperature. Even a slight change in temperature can inhibit the amplification process.
- It is complex, requiring sophisticated technology, a wide range of chemicals, and highly trained professionals to operate.
- It is not suitable for very long DNA molecules. Very long DNA needs to be cut into smaller fragments. Usually, from 0.1 kbp up to 10 kbp or 40 kbp can be used.
- RNA needs to be first converted to DNA using reverse-transcriptase enzyme before its amplification.
- Most types of PCR processes require additional steps for product analysis.
References
- Ramesh R, Munshi A, Panda SK. Polymerase chain reaction. Natl Med J India. 1992 May-Jun;5(3):115-9. PMID: 1304285.
- Staněk L. Polymerázová řetězová reakce: princip metody a využití v molekulární patologii [Polymerase chain reaction: basic principles and applications in molecular pathology]. Cesk Patol. 2013 Jun;49(3):119-21. Czech. PMID: 23964908.
- Kadri, K. (2019). Polymerase Chain Reaction (PCR): Principle and Applications. In M. L. Nagpal, O. Boldura, C. Baltă, & S. Enany (Eds.), Synthetic Biology – New Interdisciplinary Science. IntechOpen. https://doi.org/10.5772/intechopen.86491
- https://laboratoryinfo.com/polymerase-chain-reaction-pcr/
- https://www.onlinebiologynotes.com/polymerase-chain-reaction-pcr-principle-procedure-steps-types-application/
- https://microbeonline.com/polymerase-chain-reaction-pcr-steps-types-applications/
- https://www.bosterbio.com/protocol-and-troubleshooting/pcr-principle
- https://byjus.com/biology/pcr/
- Markoulatos P, Siafakas N, Moncany M. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 2002;16(1):47-51. doi: 10.1002/jcla.2058. PMID: 11835531; PMCID: PMC6808141.
- https://www.genome.gov/genetics-glossary/Polymerase-Chain-Reaction
- https://www.thermofisher.com/np/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/spotlight-articles/basic-principles-rt-qpcr.html
- https://geneticeducation.co.in/function-of-taq-dna-polymerase-in-pcr/
- (2008). PCR Primers. In: Principles and Technical Aspects of PCR Amplification. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-6241-4_5
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/biotechnology/a/polymerase-chain-reaction-pcr
- https://pediaa.com/what-is-the-difference-between-forward-and-reverse-primers/
- https://geneticeducation.co.in/what-are-the-different-components-used-in-the-pcr-reaction-buffer/
- https://www.thermofisher.com/np/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-component-considerations.html
- https://onlinelibrary.wiley.com/doi/epdf/10.1002/9781119288190.ch366
- https://geneticeducation.co.in/polymerase-chain-reaction-pcr/#PCR_requirements
- https://microbiologyinfo.com/polymerase-chain-reaction-pcr-principle-procedure-types-applications-and-animation/
- https://www.sciencedirect.com/topics/neuroscience/polymerase-chain-reaction
