Okazaki fragments are short DNA nucleotide sequences with an RNA primer at the 5′ end that are synthesized discontinuously and later joined by the enzyme DNA ligase to form the lagging strand during DNA replication.
- While researching the replication of bacteriophage DNA in Escherichia coli in 1968, Reiji Okazaki and his wife, Tsuneko Okazaki, discovered the Okazaki fragments.
- In bacteria and bacteriophage T4, Okazaki fragments range in length from 1000 to 2000 nucleotides, whereas in eukaryotes, they are approximately 150 to 200 nucleotides long.
Discovery of Okazaki fragments
- In 1963, when Tsuneko and Reiji Okazaki began studying DNA replication at Nagoya University, Japan, they concluded that semiconservative replication of DNA could be explained if daughter strands of DNA were produced in vivo by a discontinuous mechanism.
- They closely investigated the replication point to see if the daughter strand, which was developing in the 3’–5′ direction (lagging strand), may be synthesized as short fragments in the 5’–3′ direction, which is the direction opposite to the daughter DNA’s actual extension. If this were true, linking these small pieces would allow the daughter strand to lengthen.
- To verify discontinuous strand replication, Okazaki conducted a pulse-chase experiment.
- Actively replicating DNA was first combined with “hot” tritiated nucleotides for a brief pulse lasting around 5 seconds. The radioactive nucleotides were integrated into DNA strands throughout the five seconds.
- Okazaki quickly isolated the DNA by chasing after the pulse for varied durations with “cold” unlabeled nucleotides.
- After centrifuging, the DNA was checked for radioactivity. Most radioactivity was discovered in the small fragments higher in the tube following centrifuging with short chases of around 7 to 15 seconds. However, the lower, larger strands had more radiation after prolonged chases.
- This proved that small fragments on the lagging strand originate during synthesis, followed by their combination and incorporation into much bigger strands.
- These short fragments of DNA were named “Okazaki fragments” by Rollin Hotchkiss in 1968 at the Cold Spring Harbor Symposium on the Replication of DNA in Micro-organisms.
Formation of Okazaki fragments
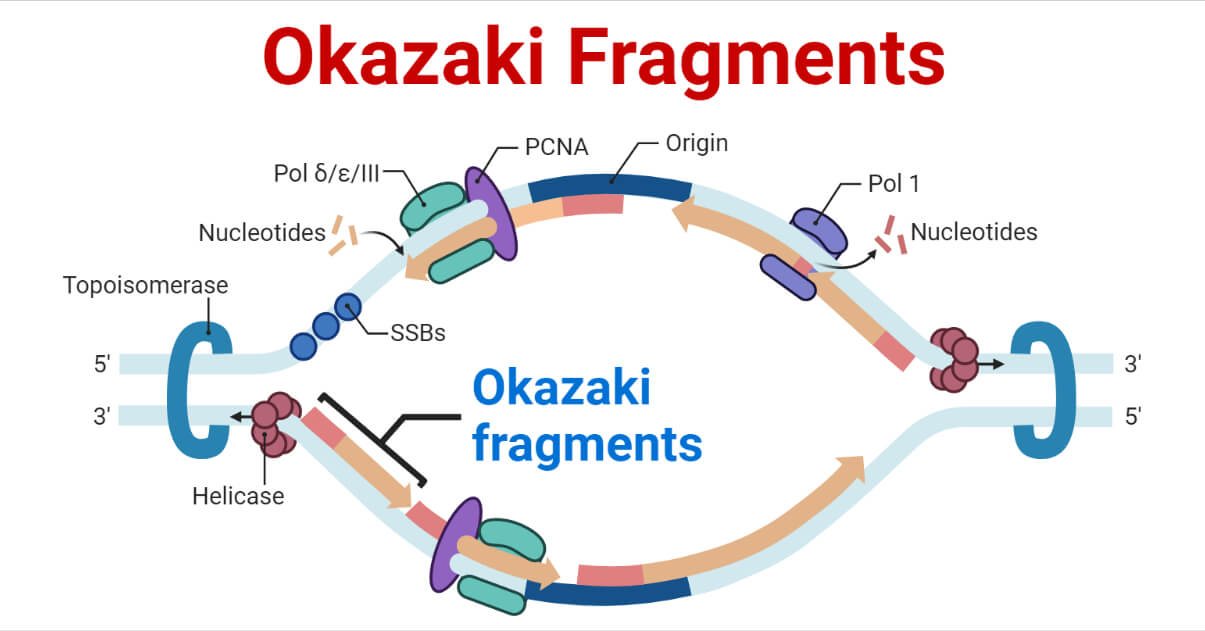
- The DNA replication fork is formed when the double helix is unwound, and the enzyme DNA helicase splits the complementary strands during DNA replication.
- Following the replication fork, DNA primase and DNA polymerase begin to act to create a new complementary strand.
- The two unwound template strands are reproduced in various ways due to the enzymes’ limited ability to operate in the 5′ to 3′ direction.
- Since the template strand of the leading strand has 3′ to 5′ directionality, DNA polymerase can assemble the leading strand while continuing to follow the replication fork. As a result, the leading strand goes through a continuous replication process.
- Due to the template strand’s 5′ to 3′ directionality, which prevents continuous strand formation, DNA polymerase must move backward from the replication fork to make the lagging strand. As a result, the lagging strand’s creation experiences periodic breaks.
- RNA primers made by primase are the starting point for synthesizing each Okazaki fragment. DNA polymerase δ in eukaryotes and DNA polymerase I in prokaryotes then extends it close to the replication fork.
- The primase and polymerase move in the opposite direction of the fork, so the enzymes must repeatedly stop and start again while the DNA helicase breaks the strands apart.
- Each new Okazaki fragment requires a new RNA primer to start the synthesis process.
- The priming occurs in three steps:
- PriA protein displaces SSB proteins from a short stretch of DNA.
- primase (DnaG) binds to PriA.
- Primase synthesizes a short RNA primer of 11-12 bases.
- The DNA Polymerase that is creating the lagging strand releases its hold on the DNA whenever a new Okazaki fragment formation begins, and it moves to a new location to begin constructing a new strand of DNA beginning at the 3′ end of the RNA primer.
- The clamp-loading complex is responsible for disassembling and moving the sliding clamp.
- Only one sliding clamp is released and reattached to a new site out of the two Polymerase III core enzymes in the replisome.
- This is so that just the lagging strand requires frequent removal and reloading of the clamp.
- The primer is later removed by enzymes that have an endonucleolytic activity, such as:
- Ribonuclease H (RNAse H)
- Flap endonucleases (FENs)
- Dna2 helicase/nucleases
FENs are distinct enzymes in eukaryotes, whereas in prokaryotes, the FEN nuclease is a domain of DNA polymerase I.
- It is yet unknown how Okazaki fragments are cleared of RNA-DNA primers.
- Using phosphodiester linkages, DNA ligase joins the fragments to form a continuous DNA strand once they have been formed.
- Given that one of the new strands forms continuously while the other does not, the complete replication process is semi-discontinuous.
Significance of Okazaki fragments
It is essential for DNA replication and cell growth that Okazaki fragments are processed well.
Okazaki fragment defects have the potential to result in strands in the DNA breaking and various chromosomal abnormalities. These chromosome mutations can change appearance, the number of sets, or the number of individual chromosomes.
References
- Balakrishnan, L., & Bambara, R. A. (2013). Okazaki fragment metabolism. Cold Spring Harbor perspectives in biology, 5(2), a010173. https://doi.org/10.1101/cshperspect.a010173
- Biology Online. (2022). Okazaki fragments. Accessed from: https://www.biologyonline.com/dictionary/Okazaki-fragment
- Clark D.P, Pazdernik N.J., & McGehee M. R. (2019). Chapter 10 – Cell Division and DNA Replication. In Molecular Biology. Third Edition. Academic Cell, pg 296-331. ISBN 9780128132883. https://doi.org/10.1016/B978-0-12-813288-3.00010-0.
- MacNeill S. A. (2001). DNA replication: partners in the Okazaki two-step. Current biology: CB, 11(20), R842–R844. https://doi.org/10.1016/s0960-9822(01)00500-0
- Okazaki T. (2017). Days weaving the lagging strand synthesis of DNA – A personal recollection of the discovery of Okazaki fragments and studies on discontinuous replication mechanism. Proceedings of the Japan Academy. Series B, Physical and biological sciences, 93(5), 322–338. https://doi.org/10.2183/pjab.93.020
- Okazaki Fragment. Accessed from: https://www.chemeurope.com/en/encyclopedia/Okazaki_fragment.html
- Professors Tsuneko and Reiji Okazaki and the Okazaki Fragment. Accessed from: https://www.itbm.nagoya-u.ac.jp/Okazaki6/Okazaki_fragment.html
- Reha-Krantz L.J. (2013). Okazaki Fragment. In Brenner’s Encyclopedia of Genetics. Second Edition. Academic Press, pg 158-160. ISBN 9780080961569. https://doi.org/10.1016/B978-0-12-374984-0.01087-1.
- Verma P.S. & Agarwal V.K. (2005). Replication of DNA. In Cell Biology, Genetics, Molecular Biology, Evolution and Ecology. Multi-color Edition. S. Chand & Company Ltd. Ram Nagar, New Delhi, pg. 31. ISBN 81-219-2442-1
