Next Generation Sequencing (NGS) is a robust platform that has enabled the sequencing of thousands to millions of DNA molecules simultaneously.
Next-generation sequencing (NGS), also known as high-throughput sequencing, is the catch-all term used to describe a number of different modern sequencing technologies.
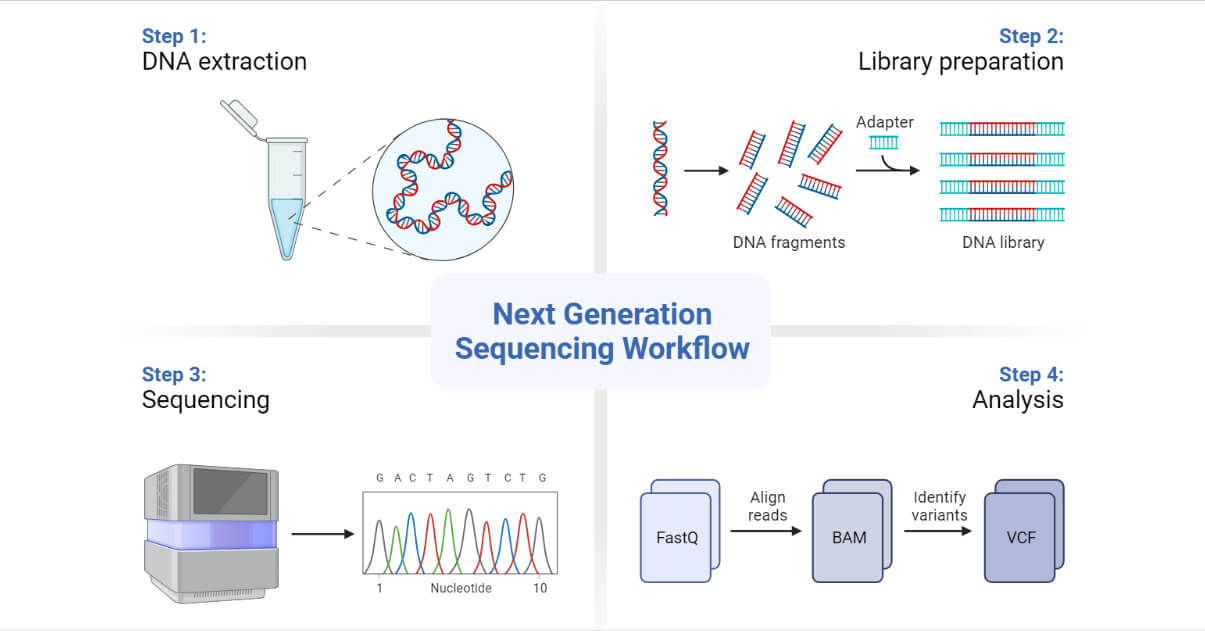
The high demand for low-cost sequencing has driven the development of high-throughput sequencing, which produces thousands or millions of sequences at once.
They are intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods.
Thus, these recent technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing and as such, have revolutionized the study of genomics and molecular biology.
Classified to different generations, NGS has led to overcoming the limitations of conventional DNA sequencing methods and has found usage in a wide range of molecular biology applications.
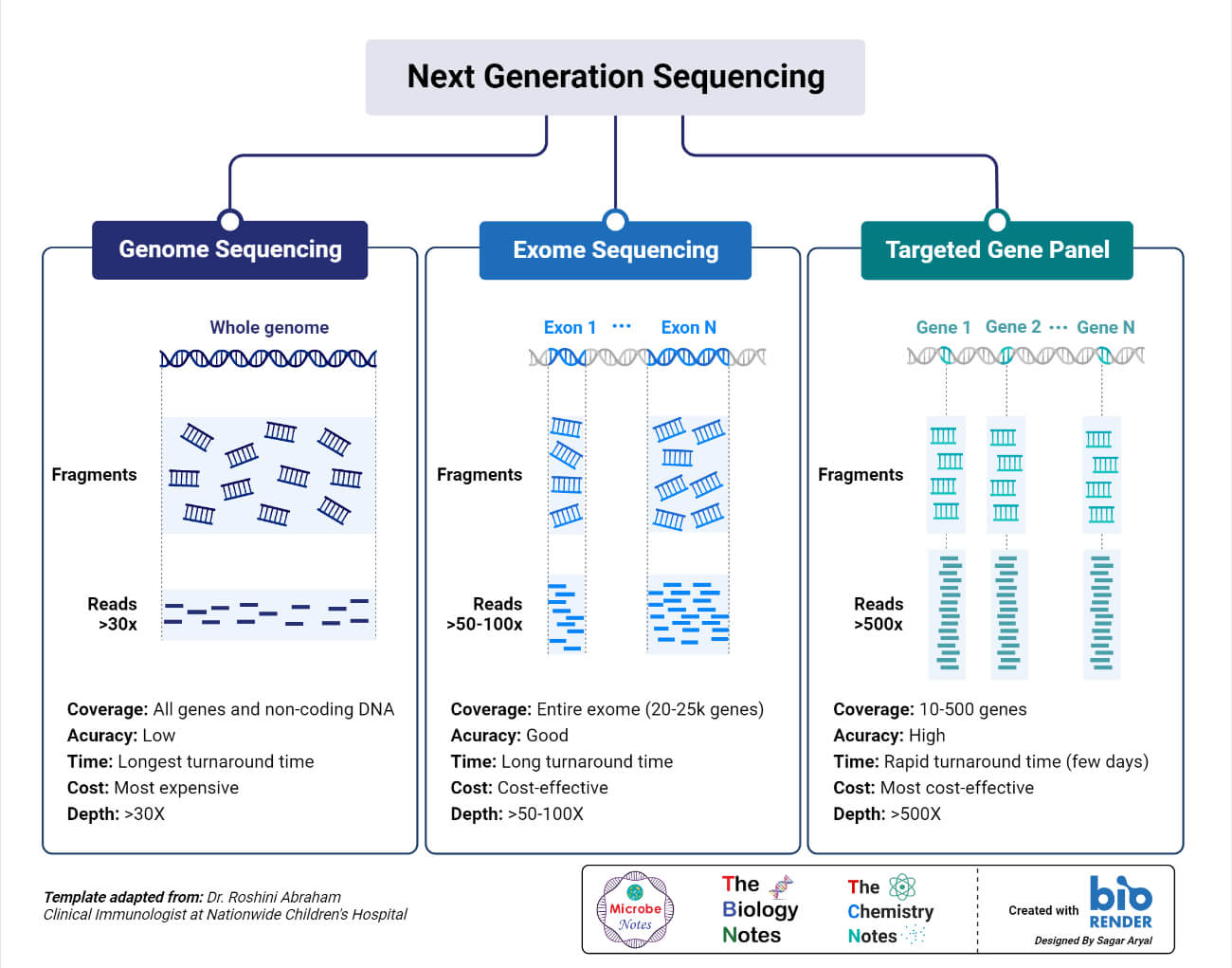
The generations it is classified into include:
First Generation
- Sanger Sequencing
Second Generation Sequencing
- Pyrosequencing
- Sequencing by Reversible Terminator Chemistry
- Sequencing by Ligation
Third Generation Sequencing
- Single Molecule Fluorescent Sequencing
- Single Molecule Real Time Sequencing
- Semiconductor Sequencing
- Nanopore Sequencing
Fourth Generation Sequencing
Aims conducting genomic analysis directly in the cell.
Interesting Science Videos
Next-Generation Sequencing Types
Lynx therapeutics’ massively parallel signature sequencing (MPSS)
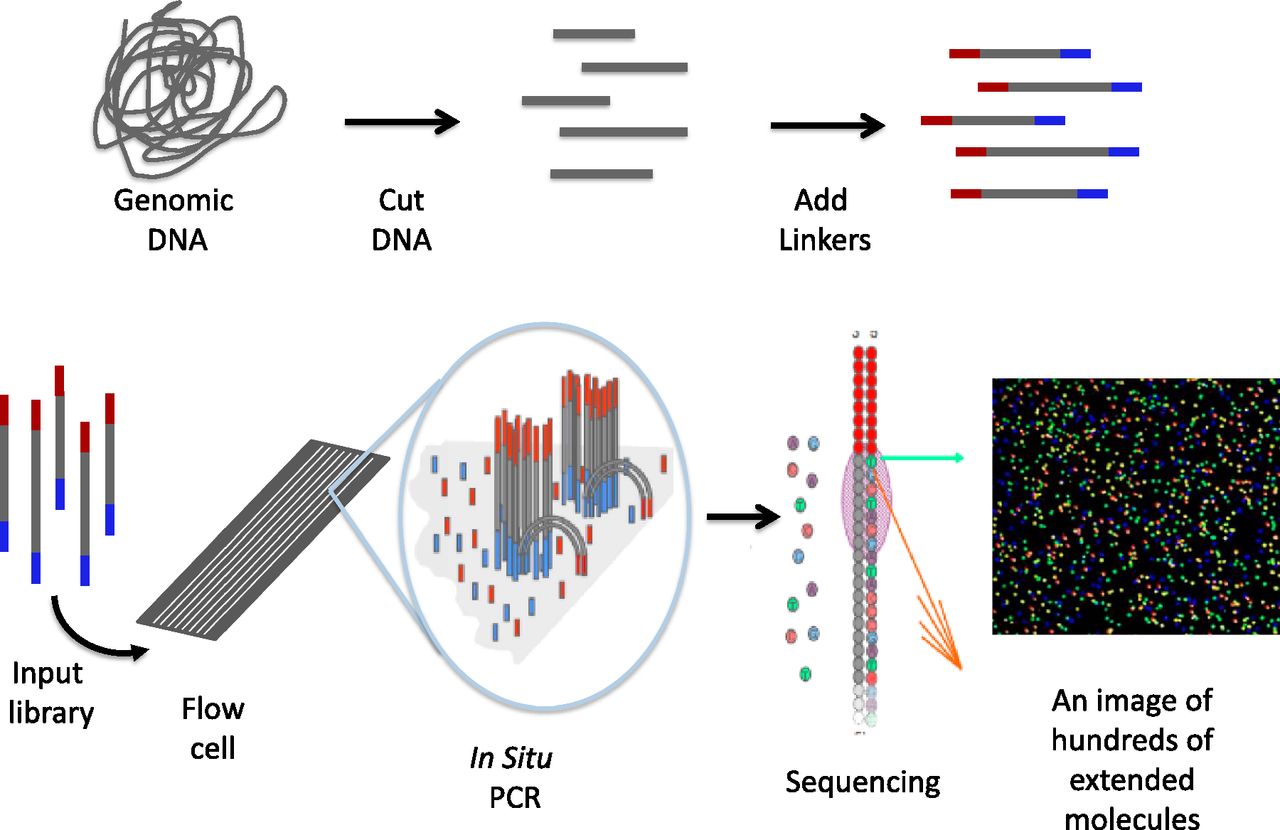
- It is considered as the first of the “next-generation” sequencing technologies.
- MPSS was developed in the 1990s at Lynx Therapeutics, a company founded in 1992 by Sydney Brenner and Sam Eletr.
- MPSS is an ultra high throughput sequencing technology.
- When applied to expression profile, it reveals almost every transcript in the sample and provide its accurate expression level.
- MPSS was a bead-based method that used a complex approach of adapter ligation followed by adapter decoding, reading the sequence in increments of four nucleotides; this method made it susceptible to sequence-specific bias or loss of specific sequences.
- However, the essential properties of the MPSS output were typical of later “next-gen” data types, including hundreds of thousands of short DNA sequences.
- In the case of MPSS, these were typically used for sequencing cDNA for measurements of gene expression levels.
Polony sequencing
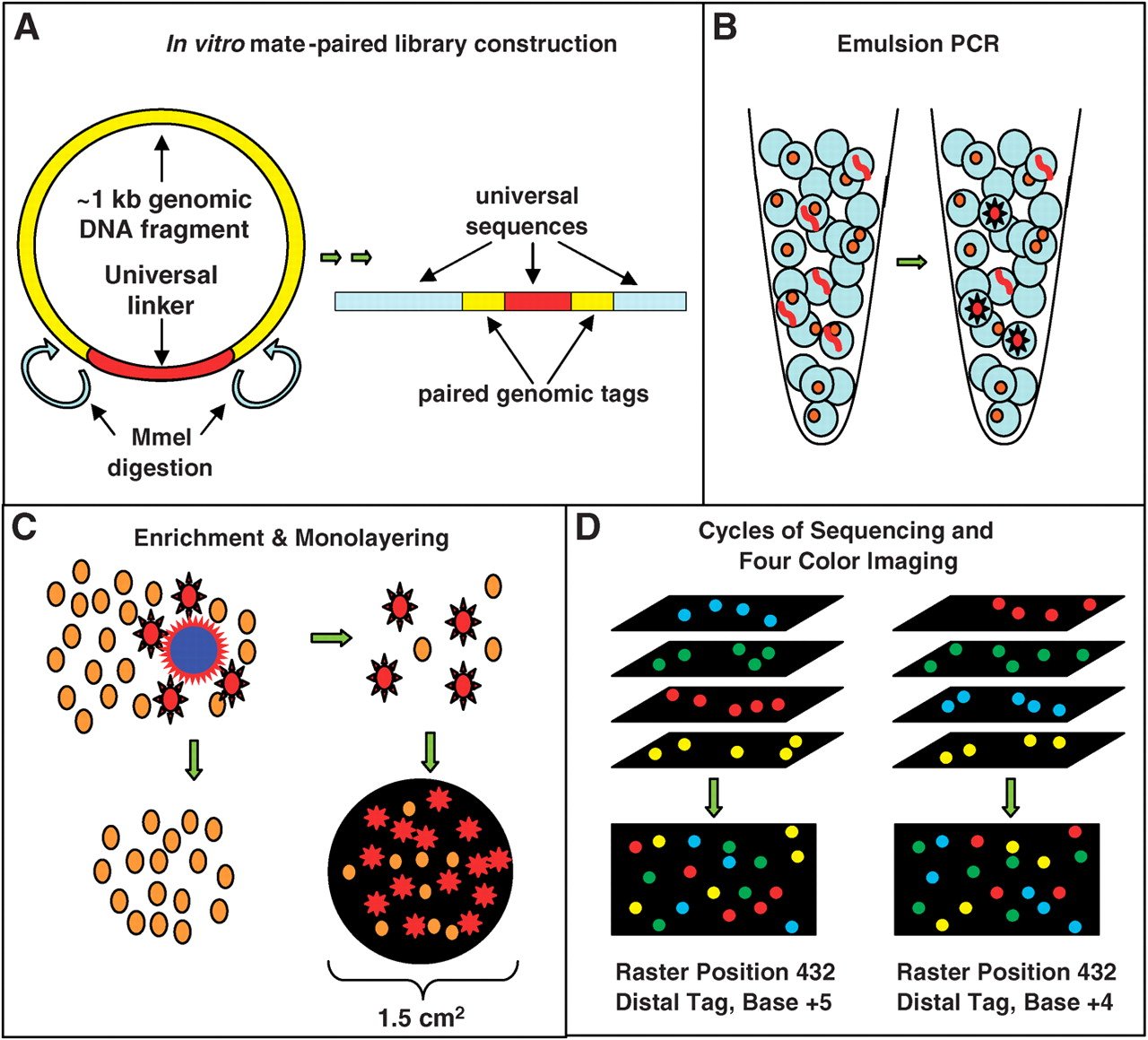
- It is an inexpensive but highly accurate multiplex sequencing technique that can be used to read millions of immobilized DNA sequences in parallel.
- This technique was first developed by Dr. George Church in Harvard Medical college.
- It combined an in vitro paired-tag library with emulsion PCR, an automated microscope, and ligation-based sequencing chemistry to sequence an E. coli genome at an accuracy of > 99.9999% and a cost approximately 1/10 that of Sanger sequencing.
Pyrosequencing
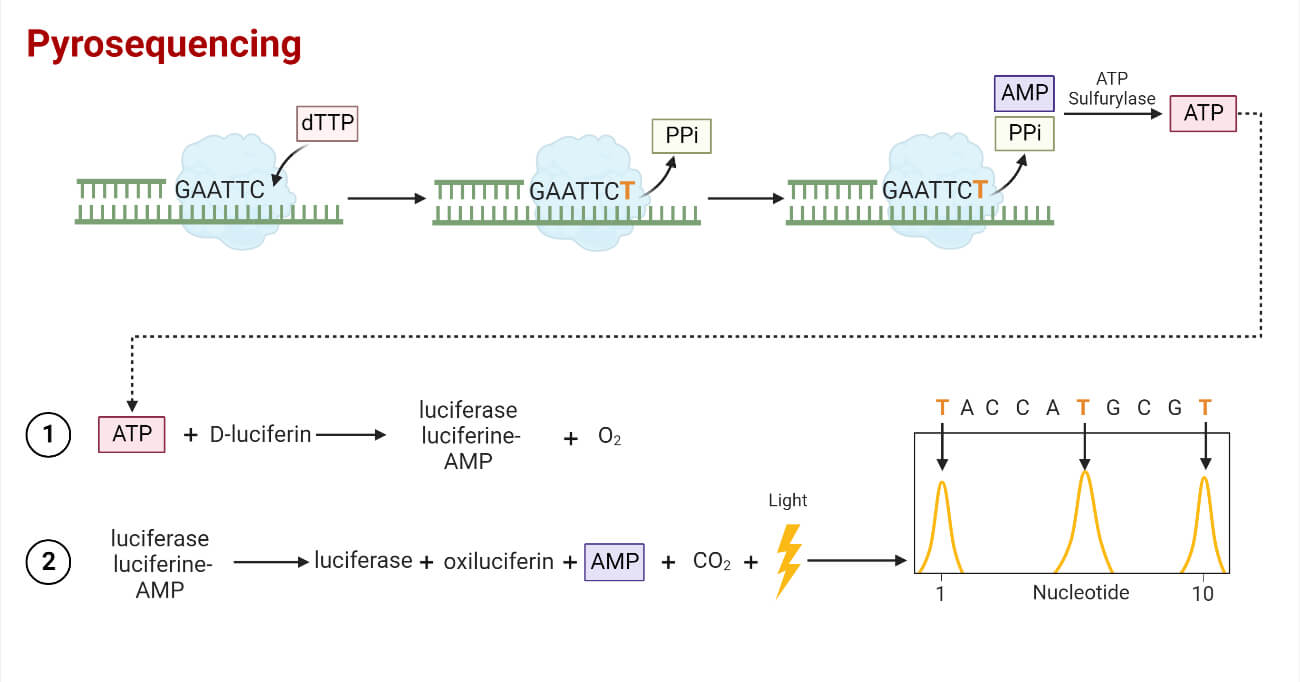
- A parallelized version of pyrosequencing was developed by 454 Life Sciences, which has since been acquired by Roche Diagnostics.
- The method amplifies DNA inside water droplets in an oil solution (emulsion PCR), with each droplet containing a single DNA template attached to a single primer-coated bead that then forms a clonal colony.
- The sequencing machine contains many picolitre-volume wells each containing a single bead and sequencing enzymes.
- Pyrosequencing uses luciferase to generate light for detection of the individual nucleotides added to the nascent DNA, and the combined data are used to generate sequence read-outs.
- This technology provides intermediate read length and price per base compared to Sanger sequencing on one end and Solexa and SOLiD on the other.
Illumina (Solexa) sequencing
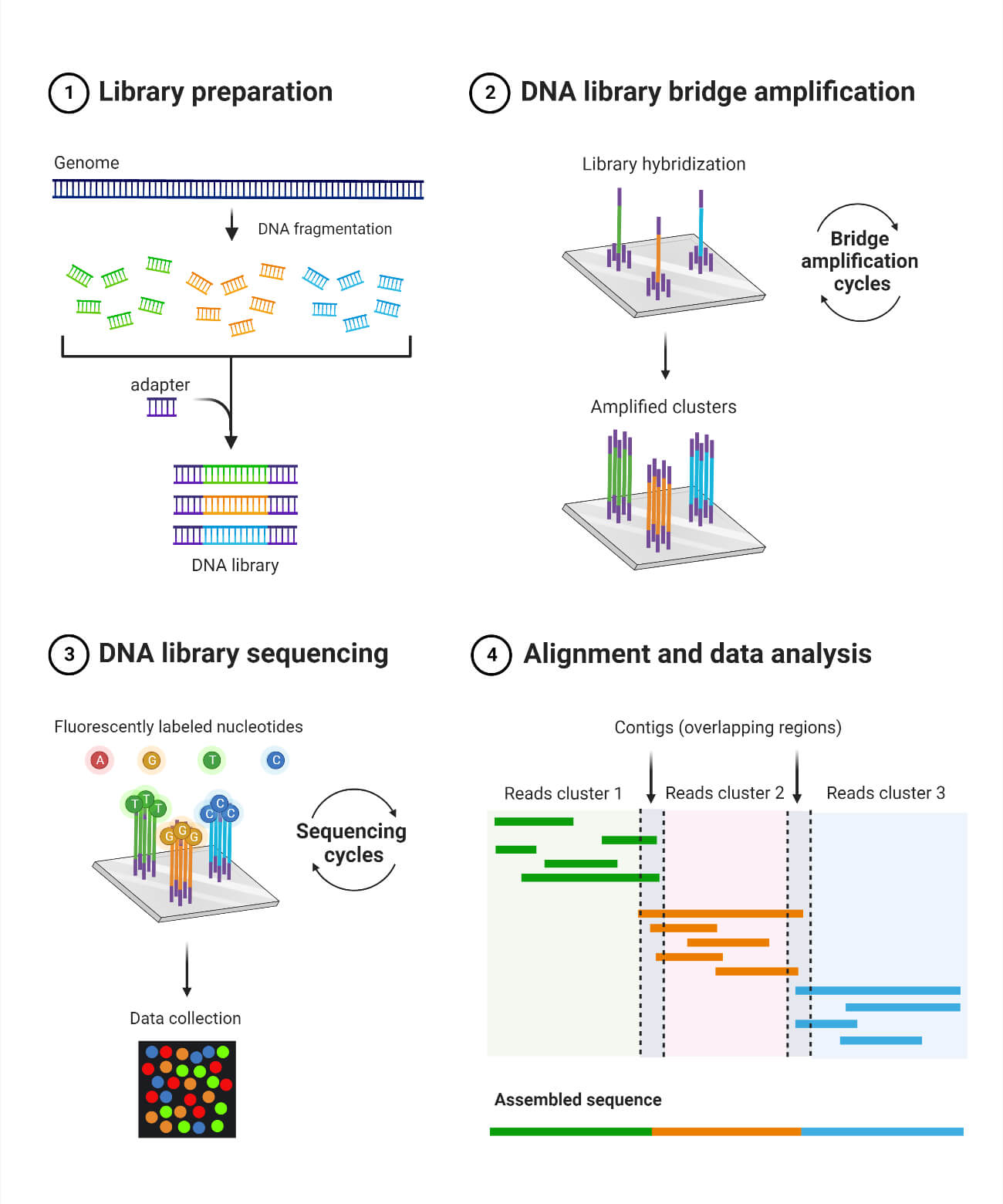
- Solexa developed a sequencing technology based on dye terminators.
- In this method, DNA molecule are first attached to primers on a slide and amplified. This is known as bridge amplification.
- Unlike pyrosequencing, the DNA can only be extended one nucleotide at a time.
- A camera takes images of the fluorescently labeled nucleotides, then the dye along with the terminal 3′ blocker is chemically removed from the DNA, allowing the next cycle to commence.
SOLiD sequencing
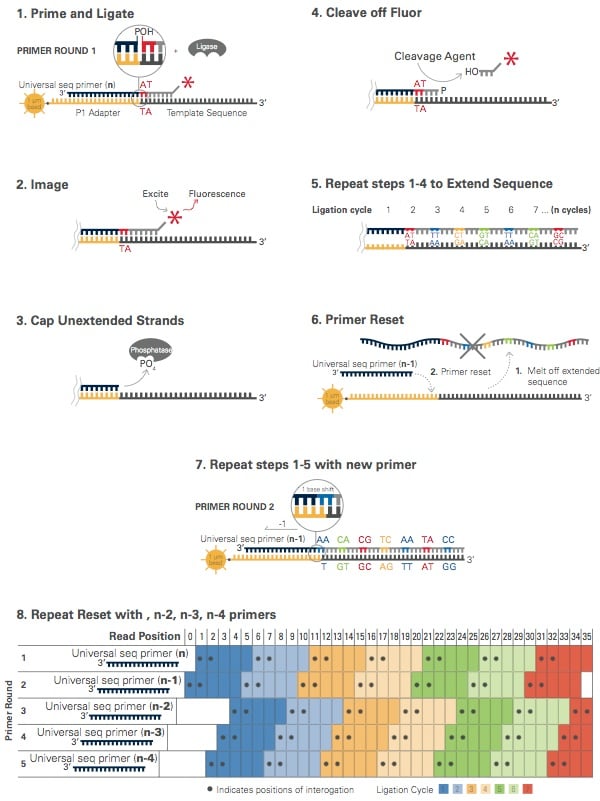
- The technology for sequencing used in ABISolid sequencing is oligonucleotide ligation and detection.
- In this, a pool of all possible oligonucleotides of fixed length are labelled according to the sequenced position.
- This sequencing results to the sequences of quantities and lengths comparable to illumine sequencing.
DNA nanoball sequencing
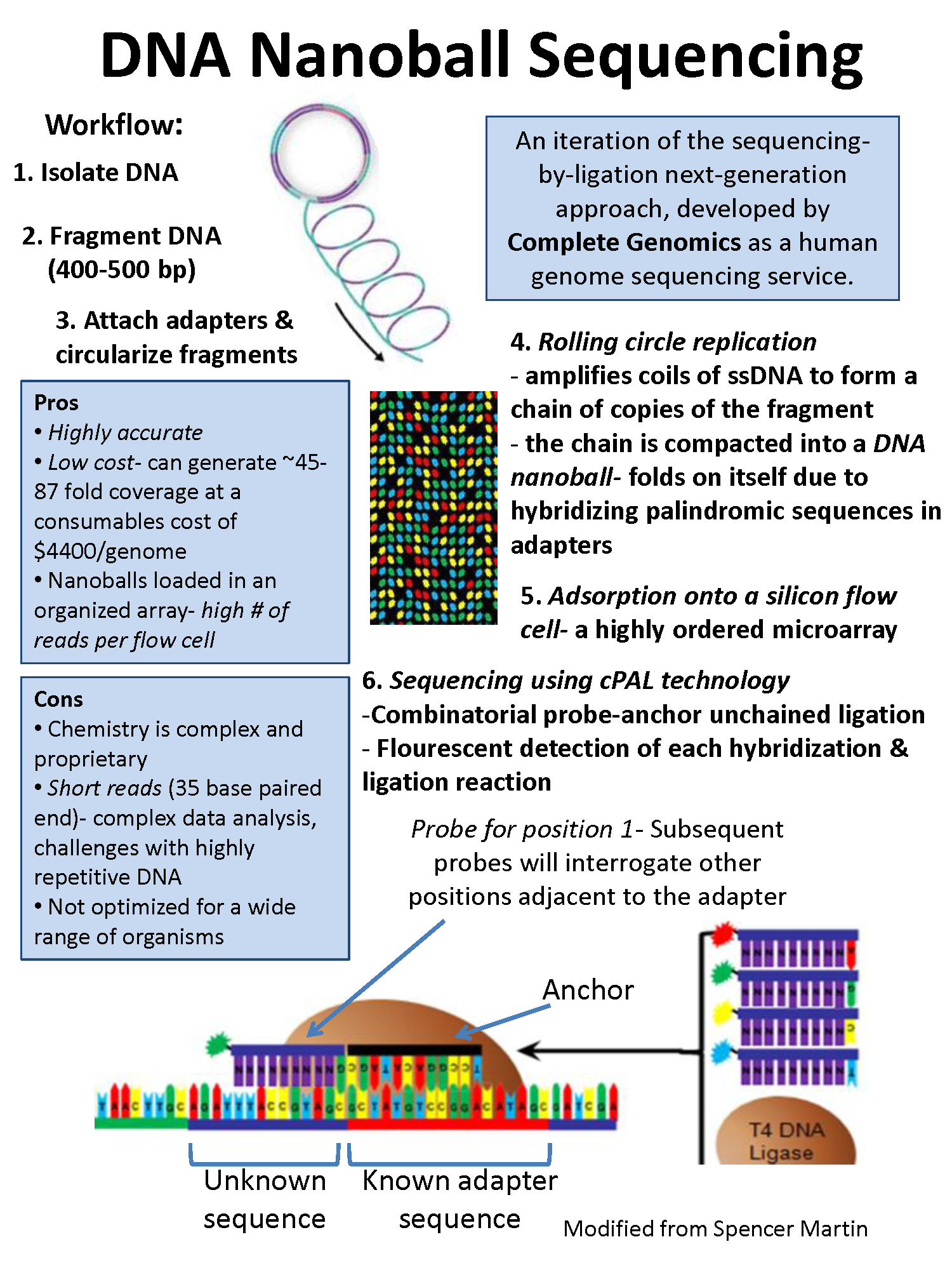
- It is high throughput sequencing technology that is used to determine the entire genomic sequence of an organism.
- The method uses rolling circle replication to amplify fragments of genomic DNA molecules.
- This DNA sequencing allows large number of DNA nanoballs to be sequenced per run and at low reagent cost compared to other next generation sequencing platforms.
- However, only short sequences of DNA are determined from each DNA nanoball which makes mapping the short reads to a reference genome difficult.
- This technology has been used for multiple genome sequencing projects and is scheduled to be used for more.
Helioscope single molecule sequencing
- Helioscope sequencing uses DNA fragments with added polyA tail adapters, which are attached to the flow cell surface.
- The next steps involve extension-based sequencing with cyclic washes of the flow cell with fluorescently labeled nucleotides.
- The reads are performed by the Helioscope sequencer.
- The reads are short, up to 55 bases per run, but recent improvement of the methodology allows more accurate reads of homopolymers and RNA sequencing.
Single molecule SMRT sequencing
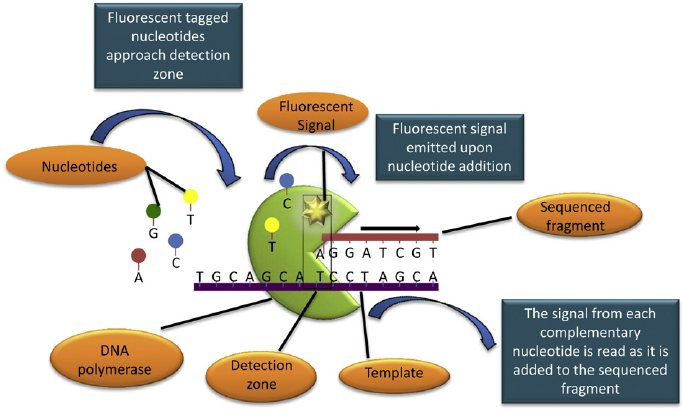
- SMRT sequencing is based on the sequencing by synthesis approach.
- The DNA is synthesisd in so called zero-mode wave-guides (ZMWs) – small well-like containers with the capturing tools located at the bottom of the well.
- The sequencing is performed with use of unmodified polymerase and fluorescently labelled nucleotides flowing freely in the solution.
- The wells are constructed in a way that only the fluorescence occurring by the bottom of the well is detected.
- The fluorescent label is detached from the nucleotide at its incorporation into the DNA strand, leaving an unmodified DNA strand.
- The SMTR technology allows detection of nucleotide modifications. This happens through the observation of polymerase kinetics.
- This approach allows reads of 1000 nucleotides.
Single molecule real time (RNAP) sequencing
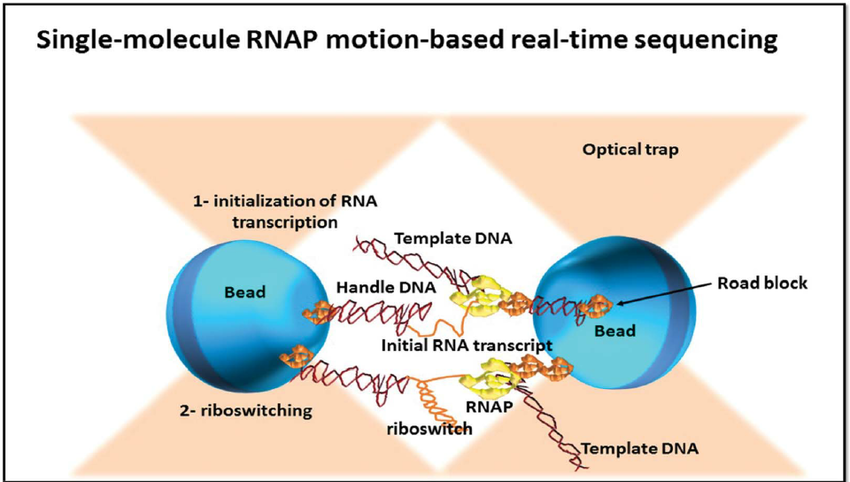
- This method is based on RNA polymerase (RNAP), which is attached to a polystyrene bead, with distal end of sequenced DNA is attached to another bead, with both beads being placed in optical traps.
- RNAP motion during transcription brings the beads in closer and their relative distance changes, which can then be recorded at a single nucleotide resolution.
- The sequence is deduced based on the four readouts with lowered concentrations of each of the four nucleotide types.
References
- http://library.umac.mo/ebooks/b28050393.pdf
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- Next-Generation Sequencing: Advantages, Disadvantages, and Future. Available from: https://www.researchgate.net/publication/306431728_NextGeneration_Sequencing_Advantages_Disadvantages_and_Future [Accessed Sep 05 2018].
- http://www.aun.edu.eg/molecular_biology/Proceeding_Dec2011/DNA%20sequencing.pdf
- https://ab.inf.uni-tuebingen.de/teaching/ws09/bioinformatics-i/10-sequencing.pdf
- http://www.pathologyoutlines.com/topic/molecularpathdnaseqmaxam.html

Thank you.
I really appreciate it because I was able to finish my homework after seeing this page you organized.
It is fascinating to read about how far DNA sequencing technology has come over the past 2 decades alone. It allows us to check for disease, find and catch criminals, and other practical uses outside of research. Thanks for explaining the differences between first, second, and third-generation sequencing. Very interesting info.