Interesting Science Videos
Major Histocompatibility Class I (MHC Class I)
- This is the first class of the MHC molecule that encodes the glycoproteins that are expressed on the surface of nearly all nucleated cells.
- Their major function is to present antigen-processed peptides to the T-cytotoxic cells by the cytosolic pathway.
- In humans, the MHC class I protein is encoded by the HLA-A, -B, and -C genes.
- This class of the MHC class I is made up of two chains i.e a transmembrane glycoprotein with a molecular weight of 45,000, which is noncovalently associated with a non–MHC-encoded polypeptide with molecular weight of 12,000 that is known as β2-microglobulin.
- Class I molecules are to be found on virtually all nucleated cells in the body except on cells in the retina and brain.
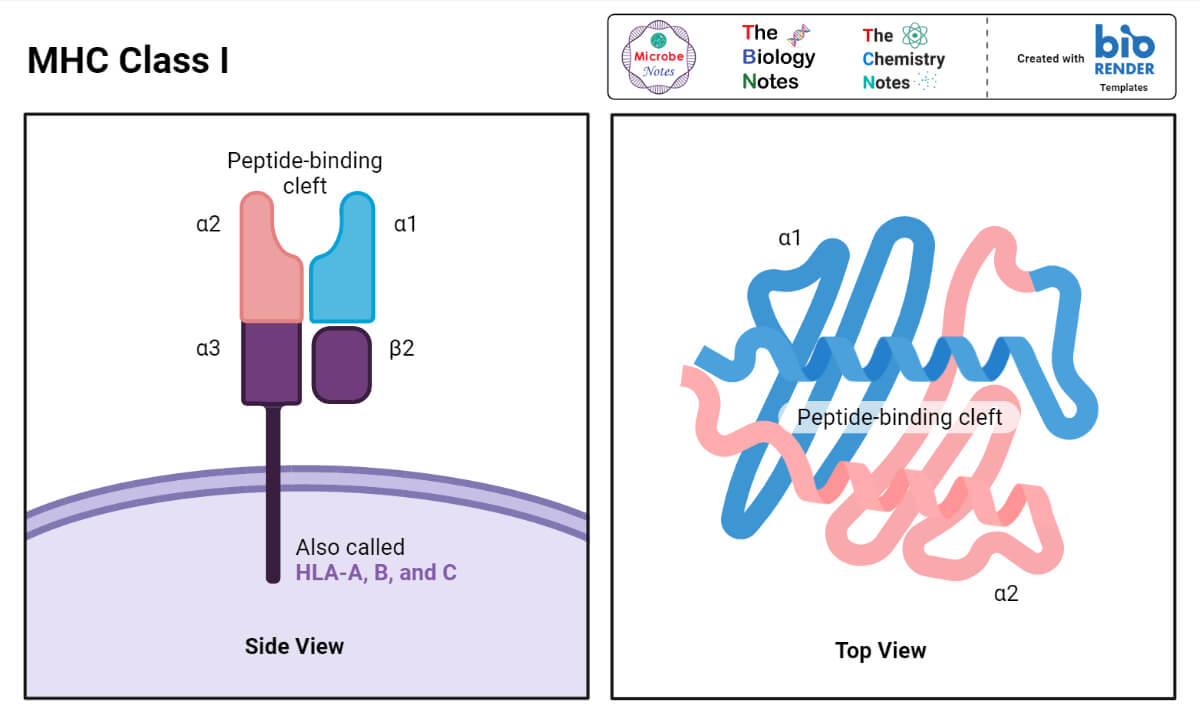
Structure of MHC Class I
- MHC Class I molecules in both human and mouse consist of two polypeptide chains that dramatically differ in size.
- The larger (α) chain has a molecular weight of 44 kDa in humans and 47 kDa in the mouse, and is encoded by an MHC Class I gene.
- The smaller chain, called β-2 microglobulin, has a molecular weight of 12 kDa in both species, and is encoded by a nonpolymorphic gene that is mapped outside of the MHC complex.
- There are no known differences in the structure of the human MHC Class I antigen a chains encoded by the HLA-A locus compared to those encoded by the HLA-B or the HLA-C loci, or in the structure of the murine MHC Class I antigen a chains encoded by the H-2K locus compared to those encoded by the H-2D or H-2L loci.
Regardless of which of these loci codes it, the α chain can be subdivided into the following regions or domains:
- the peptide-binding domain;
- the immunoglobulin-like domain;
- the transmembrane domain; and
- the cytoplasmic domain.
- The peptide-binding domain is the most N-terminal; it is the only region of the molecule where allelic differences in the amino acid sequence can be localized.
- As seen from its name, the peptide-binding domain of the molecule includes the site to which antigenic peptides bind.
- It makes much sense to have this site exactly where the allelic differences are, because different MHC alleles accommodate peptides better or worse, thus influencing on the magnitude of the T-cell response.
- X-ray crystallography showed that the peptide-binding site in the MHC Class I molecules looks like a cleft that has a ‘‘floor’’ and two ‘‘walls’’ formed by spiral shaped portions of the alpha chain, called alpha 1 and alpha 2.
- Since the ‘‘floor’’ of the peptide-accommodating cleft is closed, only relatively small peptides, consisting of 9 to 11 amino acid residues, can be ‘‘stuffed’’ there.
- The immunoglobulin-like domain is structurally conserved, and resembles a domain of an antibody C-region.
- It contains the binding site for the T-cell accessory molecule CD8.
- The transmembrane and the cytoplasmic domains ensure that the alpha chain spans the membrane and is properly expressed by the cell.
- The β-2-microglobulin chain is also vitally important for the proper expression of the alpha chain.
- There are some mutant lymphoid cell lines (notably Daudi) that do not express MHC Class I molecules because of the defect in the β-2-microglobulin gene.
Major Histocompatibility Class II (MHC Class II)
- The class II MHC genes encode glycoproteins expressed primarily on antigen-presenting cells (macrophages, dendritic cells, and B cells), where they present processed antigenic peptides to TH cells.
- The class II proteins are encoded by the HLA-D region and the HLA-D regions have three families, DP-, DQ-, and DR-encoded molecules.
- This class retains control of immune responsiveness and the different allelic forms of these genes confer differences in the ability to mount an immune response against a given antigen.
- The HLA-D locus-encoded proteins are made up of two noncovalently associates transmembrane glycoproteins with a molecular weight of 33,000 and 29,000 respectively.
- They have a restricted tissue distribution and they are chiefly found on macrophages, dendritic cells, B-cells, and other antigen-presenting cells. They are also expressed on other cells such as endothelial cells and/or epithelial cells induced by IFN-γ.
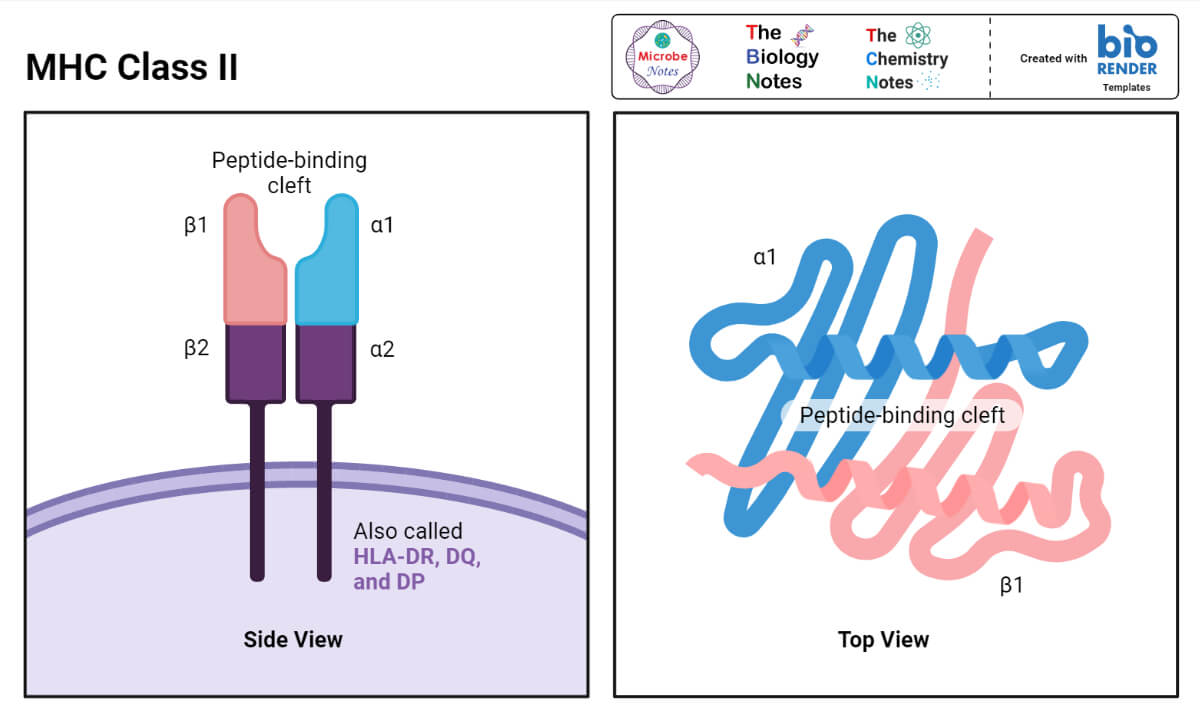
Structure of MHC Class II
- Class II MHC molecules in both humans and mice consist of two polypeptide chains that have a similar, albeit not identical size.
- One of them is called alpha (α) and the other beta (β).
- The molecular weight of the α chain is 32–34 kDa, and of the β chain 29–32 kDa.
- A separate gene controls each of the chains.
- Thus, the murine I-A locus actually consists of the Iα and Iβ genes, the human HLA-DR locus of the HLA-DRα and HLA-DRβ, etc. Both the α and the β genes are polymorphic.
- The β genes of some of the MHC Class II loci can be tandemly duplicated, so, instead of one gene per homologous chromosome, a cell can have two or three.
- Because of that, one cell can simultaneously express more than two allelic products of each of the MHC Class II loci.
- For example, a cell can express allelic products of its HLA-DR molecule that can be identified as HLADRα1– HLA-DRβ1; HLA-DRα2 – HLA-DRβ2; HLA-DRα1 – HLA-DRβ2; HLA-DRα2 – HLA-DRβ1; etc.
- Overall, one cell can simultaneously express as many as 20 different MHC Class II gene products because of this tandem duplication phenomenon.
The structure of the α and the β chains of the MHC Class II molecules resembles that of the alpha chain of the MHC Class I molecules in that the former can be also divided into the peptide-binding, the immunoglobulin-like, the transmembrane, and the cytoplasmic domains.
One important difference, however, is that the peptide-binding cleft in Class II molecules is formed by both alpha and beta chains.
Although positioned close to each other in space, the spirals of the alpha and the beta chains that form the cleft are not physically bound to each other.
Because of that, the ‘‘floor’’ of the peptide-accommodating cleft in Class II MHC molecules is ‘‘open,’’ or ‘‘has a hole’’ in it.
That allows MHC Class II molecules to accommodate peptides that are larger than those that fit MHC Class I molecules.
The immunoglobulin-like domain of the MHC Class II molecules contains the binding site for a T-cell accessory molecule, CD4.
This site cannot bind the above-mentioned CD8 molecule.
Major Histocompatibility Class III (MHC Class III)
- Class III MHC genes encode for various secreted proteins that have immune functions, including the component of the complement system and molecules that are involved in inflammation such as cytokines.
Antigen Processing and Presentation
- The recognition of protein antigens by T-lymphocytes required that the antigens be processed by Antigen-presenting Cells, then displayed within the cleft of the MHC molecules on the membrane of the cell.
- This involves the degradation of the protein antigens into peptides, a process known as antigen processing.
- When the antigen has been processed and degraded into peptides, it then associates with MHC molecules within the cell cytoplasm forming a peptide-MHC complex. This complex is then transported to the membrane, where it is displayed by a process of antigen presentation.
- The MHC Class I and class II MHC molecules associated with peptides that have been processed in different intracellular compartments.
- The Class I MHC molecules bind peptides derived from endogenous antigens that have been processed within the cytoplasm of the cell such as tumor proteins, bacterial proteins, or viral proteins, or cellular proteins, and processed within the cytosolic pathway.
- Class II MHC molecules bind peptides derived from exogenous antigens that are internalized by phagocytosis or endocytosis and processed within the endocytic pathway.
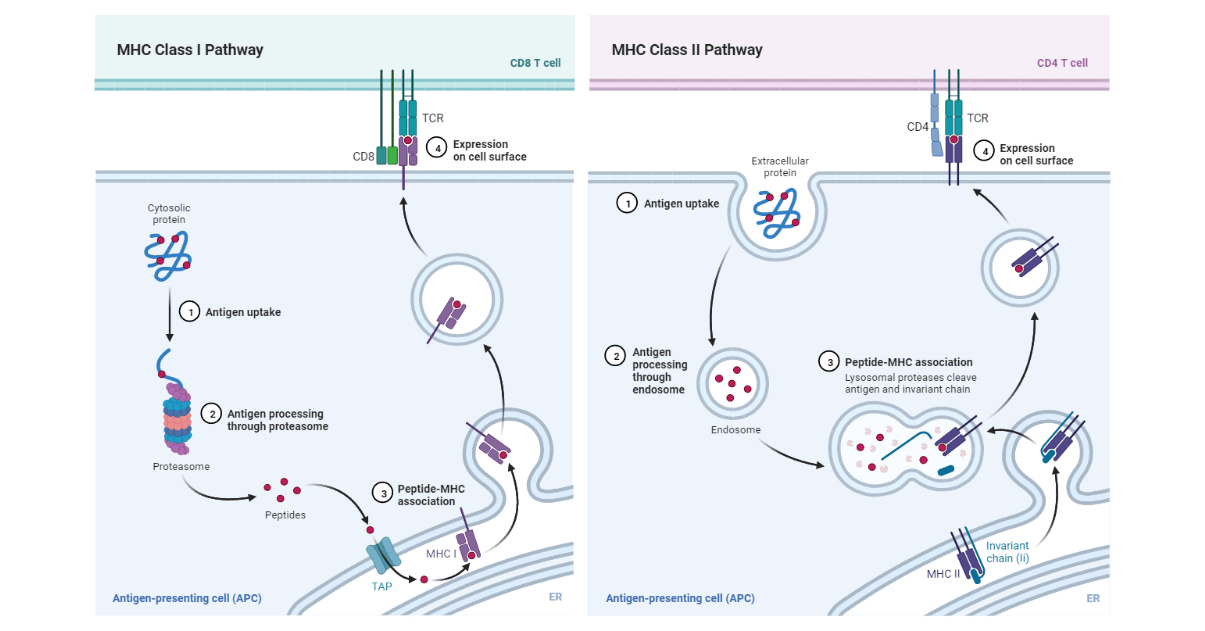
A. Cytosolic pathway: Endogenous antigen
- This is the pathway that processes and presents the endogenous antigen using the Class I MHC molecules.
- The antigen proteins are degraded intracellularly to short peptides by a cytosolic proteolytic system that is present in all cells. These proteins targeted for proteolysis have a small protein known as ubiquitin attached to them.
- The ubiquitin-protein conjugate then gets degraded by a multifunctional protease complex known as a proteasome.
- Each proteasome is a large (26S), cylindrical particle that consists of four rings of protein subunits and a central channel of 10–50 Å diameter.
- The proteasome can cleave peptide bonds between 2-3 different amino acid combinations in an ATP-dependent process.
- Degradation of the ubiquitin-protein complex takes place in the central hollow of the proteasome.
- The peptides are then transported from the cytosol to the rough endoplasmic reticulum. This is enabled by the transporter protein, designated TAP (transporter associated with antigen processing) is a membrane-spanning heterodimer consisting of two proteins: TAP1 and TAP2.
- The TAP1 and TAP2 proteins each have a domain projecting into the lumen of the Rough endoplasmic reticulum (RER), and an ATP-binding domain that extends into the cytosol.
- Both TAP1 and TAP2 belong to the family of ATP-binding cassette proteins found in the membranes of many cells, including bacteria.
- They mediate ATP-dependent transport of amino acids, sugars, ions, and peptides.
- the peptides that are generated in the cytosol by the proteasome, are translocated into the Rough Endoplasmic Reticulum (RER) by TAP proteins by a process that utilizes hydrolyzed ATP. TAP proteins have a high affinity for peptides sizes of 8-10 amino acids, the optimum length for class I MHC binding.
- Additionally, TAP proteins favor peptides with hydrophobic or basic carboxyl-terminal amino acids, which is the preferred anchor residue for class I MHC molecule, and therefore, TAP is optimized to transport peptides that will interact with class I MHC molecules.
- Next, the peptides that are assembled with class I MHC are aided by chaperone molecules that facilitate the folding of polypeptides.
- The alpha and beta-2-microglobulin components of the class I MHC molecules are synthesized on the polysomes along the rough endoplasmic reticulum. These components are assembled into a stable class I MHC molecules complex that can exit the RER requiring the presence of a peptide in the binding groove of the class molecule.
- The first chaperone involved is known as calnexin, which is a resident membrane protein of the endoplasmic reticulum. Calnexin associates with the class I α chain and promotes its folding. When the Beta-2-microglobulin binds to the α chain, the calnexin is released, and the class I molecule associates with the chaperone calreticulin and with tapasin.
- Tapasin is a TAP-associated protein that brings the TAP transporter into proximity with the class I molecule and allows it to acquire an antigenic peptide. The physical association of the α chain-beta-2-microglobulin heterodimer with the TAP protein promotes peptide capture by the class I molecule before the peptides are exposed to the RER.
- The peptides not bound by class I molecules are rapidly degraded.
- After binding, the class I molecule displays increased stability and can dissociate from calreticulin and tapasin, exit from the RER, and proceed to the cell surface via the Golgi.
- An additional chaperone protein, ERp57, associates with calnexin and calreticulin complexes. The precise role of this resident endoplasmic reticulum protein in the class I peptide assembly and loading process has not yet been defined, but it is thought to contribute to the formation of disulfide bonds during the maturation of class I chains.
B. Endocytic Pathway: Exogenous antigen
Antigen-presenting cells can internalize antigen by phagocytosis, endocytosis, or both. Macrophages internalize antigen by both phagocytosis and endocytosis. Most of the other APCs are poorly phagocytic and can only internalize the antigen by pinocytosis or endocytosis, whereas most other APCs are not phagocytic and therefore they internalize the exogenous antigen only by endocytosis of by pinocytosis. B-cells which are also APCs internalizes the antigen effectively by receptor-mediated endocytosis using antigen-specific membrane antibody receptors.
- When the exogenous antigen is internalized, it is degraded into peptides in the compartments of the endocytic processing pathway.
- The breaking down of antigens into peptides takes 1-3 hours to transverse the endocytic pathway and appear at the cell surface in the form of a peptide-class II MHC complex.
- In this pathway, three acidic compartments: early endosome (pH 6.0-6.5), late endosome or endolysosomes (pH 5.0-6.0); and lysosomes (pH 4.5-5.0). The internalized antigen moves from the early to late endosomes and later to the lysosomes where they encounter the hydrolytic enzyme, with a decreasing pH in each compartment.
- the lysosomes have a unique collection of 40 acid-dependent hydrolases including proteases, nucleases, glycosidases, lipases, phospholipases, and phosphatases. Within the compartments of the endocytic pathway, the antigen is degraded into oligopeptide made up of 13-18 residues, that bind to class II MHC molecule. The hydrolytic enzymes are active in low Ph, they inhibit antigen processing chemical agents that may increase the compartment pH and that of protease inhibitors.
- Movement of the peptides from one compartment to the next has been associated with small transport vesicles.
- After getting to the final compartments, they return to the cell periphery fusing with the plasma membrane, enabling the recycling of surface receptors.
- The antigen-presenting cells express both MHCI and MHC II molecules, therefore to prevent binding of MHC II to the same set of antigenic peptides as those of class I MHC, some mechanisms must exist to prevent this.
- When the MHC II has been synthesized within the RER, three pairs of class II chains associate with a preassembled trimer of a protein known as an invariant chain (Ii, CD74). The trimeric protein interacts with the peptide-binding cleft of the class II MHC molecules, preventing any endogenously derived peptides from binding to the cleft while the MHC class II remains within the RER.
- The invariant chain is also involved in the folding of class II MHC and its chains, the exit from the RER, and routing it to the endocytic processing pathway from the trans-Golgi network into the endocytic vesicles.
- Secondly, the peptides assemble with class II MHC molecule by displacing CLIP (Class-II associated invariant chain peptide). Most of class II MHC-invariant chain complexes are transported from the RER where they are formed through the Golgi complex and trans-Golgi network, and then through the endocytic pathway, moving from early endosomes to late endosomes then finally to the lysosomes.
- This increases the proteolytic activity from each compartment to the next.
- This causes the degradation of the invariant chain gradually, leaving a short fragment of the invariant chain known as the CLIP (Class II-associated invariant chain peptide) that remains bound to the class II molecule after the invariant chain has been cleaved with the endosomal compartment.
- CLIP occupies the peptide-binding groove of the class II MHC molecule, preventing premature binding of the antigenic peptide. HLA-DM molecule catalyzes the exchange of CLIP with the antigenic peptides. It is found in mammalian cells, mice, and rabbits. HLA-DM is neoclassical and nonpolymorphic.
- When the HLA-DM and class II CLIP complex react, it facilitates the exchange of CLIP for another peptide but in the presence of HLA-DO, it can bind to HLA-DM reducing the efficiency of the exchange reaction.
- The HLA-DO which has a similar structure as that of HLA-DM helps to modulate the function of HLA-DM, however, the function is obscure.
Presentation of Non-peptide antigens
- Nonpeptide antigens are also recognized by the immune system, these are antigens that are derived from infectious agents such as Mycobacterium tuberculosis.
- These antigens are recognized by T-cell Receptors known as δγ-TCR (T-cell receptor are dimers of αβ and δγ) which are derived from glycolipid of bacterial pathogens such as Mycobacterium tuberculosis.
- These nonprotein antigens are presented by members of the CD1 family of nonclassical class I molecules.
- The CD1 family of molecules associates with β2-microglobulin and it has its structure similar to that of MHC I molecules. It has 5 genes that encode for human CD1 molecules (CD1A-E, encoding the gene products CD1a-d, no E has been identified yet. These genes are located on the chromosomes and not on MHC I.
- They are classified into two groups based on sequence homology. Group 1 includes CD1A, B, C, and E; CD1D is in group 2. All mammalian species have CD1 genes, although the number varies. Rodents have only group 2 CD1 genes, whereas rabbits, like humans, have five genes, including both group 1 and 2 types.
- The sequence identity of CD1 with classical class I molecules is considerably lower than the identity of the class I molecules with each other. CD1D1 as compared to class I MHC shows that the antigen-binding groove of Cd1d1 is deeper and more voluminous than that of class I MHC molecule.
Clinical Significance of Antigen processing and presentation
- Sometimes the antigen-presenting cells (APCs) can deliver self-antigens which cause autoimmune diseases. When the self-antigens are presented to the T-cells, it initiates an immune reaction against our own tissues, causing autoimmune disorders such as Graves Disease, rheumatoid arthritis.
- In Graves’ disease, TSHR (Thyroid-stimulating hormone receptors) acts as the self-antigen, which is presented to T-cells activating B-cells which produce autoantibodies against TSHRs in the thyroid. This leads to the activation of TSHRs causing hyperthyroidism and leading to goiter.
References
- Immunology by Kuby, 5th Edition
- Microbiology by Prescott, 5th Edition
- Lippincott’s Illustration Review in Immunology
- Comprehensive immunology: Antigens and Immunogens by Frederic C. McDuffie
- Medical Microbiology by Jawertz, 23rd Edition
- Immunobiology: The Immune System in Health and Disease. 5th edition: Chapter 3, Antigen Recognition by B-cell and T-cell Receptors
- http://resources.schoolscience.co.uk/unilever/16-18/proteins/Protch7pg1.html
- https://microbiologyonline.org/index.php/about-microbiology/microbes-and-the-human-body/antibody-antigen-complex
- https://www.sigmaaldrich.com/technical-documents/articles/biology/antigens-epitopes-antibodies.html
- https://www.technologynetworks.com/immunology/articles/antigen-vs-antibody-what-are-the-differences-293550
- https://en.wikipedia.org/wiki/Antigen
- https://www.britannica.com/science/antigen
- https://www.webmd.com/diabetes/qa/what-are-antigens
- https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Kaiser)/Unit_6%3A_Adaptive_Immunity/12%3A_Introduction_to_Adaptive_Immunity/12.2%3A_Antigens_and_Epitopes
- https://www.news-medical.net/life-sciences/The-Characteristics-of-Antigens.aspx
- https://www.biologyonline.com/dictionary/tolerogen
- https://www.toleranzia.com/technology/tolerogen-platform/
- https://en.wikipedia.org/wiki/Tumor_antigen
- https://www.prospecbio.com/viral_antigens
- https://www.creative-diagnostics.com/Viral-Antigens.htm
- https://www.msdmanuals.com/professional/hematology-and-oncology/tumor-immunology/tumor-antigens
- https://en.wikipedia.org/wiki/Cluster_of_differentiation
- https://www.sinobiological.com/research/cd-antigens/what-is-cluster-of-differentiation
