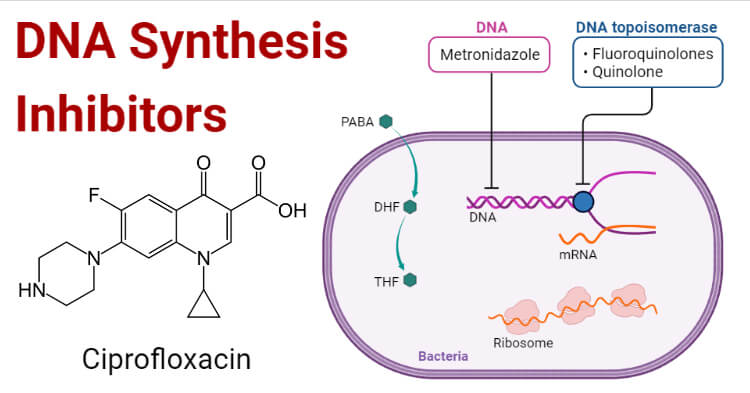
What are DNA Synthesis Inhibitors?
DNA synthesis occurs at chromosomes which requires substrates, primers, templates, and enzymes. Different drugs were used in treating diseases by inhibiting DNA synthesis in different manners. Analogs of purine and Pyrimidine’s which are used in oncology and Fluoroquinolones which are used in treating bacterial infections that inhibit topoisomerase II and IV and DNA gyrase enzyme that is used in DNA replication. The enzymes topoisomerases and gyrase have a quite similar structure but they operate differently. Gyrase removes stress while topoisomerases play a major role in removing knots that occurs in the bacterial chromosome. Subunits of gyrase are GyrA and GyrB whereas subunits of topoisomerase in Gram-positive is GrlA and GrlB and ParC and ParB. These subunits contain different domains that are involved in DNA cleavage. Humans also share two types of topoisomerase enzymes that have the same type of amino acids which is creating an issue in generating quinolones.
Fluoroquinolones are the group of antibiotics that are used in treating infections caused by both Gram-positive and Gram-negative organisms. Nalidixic acid which was introduced in 1962 was the first quinolone for clinical use. Based on their activity, quinolones are divided into different generations. They are bicyclic core structure which includes carboxyl acid group and carbonyl at position 3 and 4 respectively which enhances the activity of quinolones. Quinolones are divided into three categories which include pyrrolidinyl, piperazinyl, and piperidinyl. Piperazinyl type shows wide activity against Gram-negative organisms while Pyrrolidinyl and Piperidinyl show wide activity against both types of bacteria.
In 1977, gyrase was identified as the target for quinolones and later it was confirmed that it was a primary target for quinolones in Gram-positive bacteria.
Mechanism of action of DNA Synthesis Inhibitors
For cell survival topoisomerase and gyrase generate breaks in chromosomes. Thus, these enzymes can fragment the genome. Quinolones increase the concentrations of these enzymes which result in causing cell death. Quinolones convert gyrase and topoisomerase into toxins so they are referred to as topoisomerase poison. In a non-covalent manner, quinolones bind in the cleavage-ligation active site and interact with the proteins causing intercalation into the DNA. As a result of intercalation, the concentration of cleavage complexes increases that block ligation.
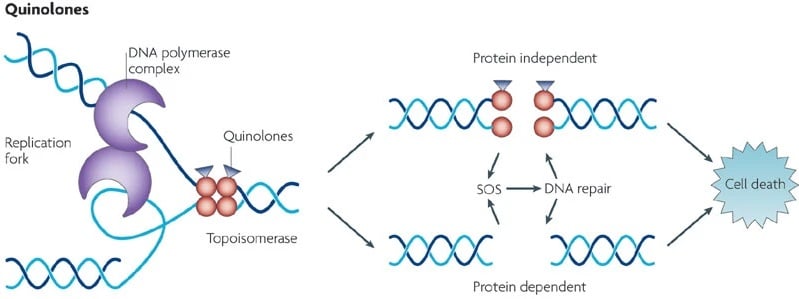
Then, after the collision of drug stabilized gyrase or topoisomerase with transcription complexes and replication forks, DNA cleavage complexes are changed to chromosomal breaks. Because of these DNA breaks, SOS response and DNA repair pathways are affected causing cell death. As quinolones inhibit DNA ligation by stabilizing cleavage complexes and also affect the functions of both enzymes. So, quinolones also act as catalytic inhibitors.
Examples of DNA Synthesis Inhibitors
- Ciprofloxacin
- Levofloxacin
- Moxifloxacin
- Ofloxacin
Mechanism of resistance of DNA Synthesis Inhibitors
Mutations in enzymes may cause quinolone resistance. Genes that cause a chromosomal mutation that codes for protein targets and plasmid-mediated genes. Mutation of serine or acidic residue decreases the affinity of drug and enzyme complex. And disruption of the metal ion bridge may also cause quinolone resistance. Mutations at acidic residue decrease catalytic activity while serine residue in gyrase and topoisomerase IV does not affect catalytic activity.
Plasmid-mediated quinolone resistance: Qnr genes that encode proteins and McbG and MfpA decrease the binding of enzymes to DNA. Thus, quinolones were protected and these genes bind to gyrase and topoisomerase and inhibit quinolones from entering into complexes.
Chromosome mediated resistance: Expression of porins facilitates drug efflux which leads to resistance.
References
- Aldred K J, Kerns R J and Oshero N (2014). Mechanism of Quinolone Action and Resistance.
- Cozzarelli N R (1977). The mechanism of action +958 of inhibitors of DNA synthesis. (c).
- Fàbrega A, Madurga S, Giralt E and Vila J(2009). Review Mechanism of action of and resistance to quinolones. 2: 40–61. https://doi.org/10.1111/j.1751-7915.2008.00063.x
- From, http://www2.csudh.edu/nsturm/CHEMXL153/DNASynthesis.html
- Kohanski, M., Dwyer, D. & Collins, J. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8, 423–435 (2010). https://doi.org/10.1038/nrmicro2333.
