DNA replication is the process of producing two identical copies of DNA from one original DNA molecule.
- DNA is made up of millions of nucleotides, which are composed of deoxyribose sugar, with phosphate and a base.
- The complementary pairing of these bases keeps the double strands intact. So, to make two copies of one DNA, these hydrogen bonds in between the bases should be broken to begin replication.
- DNA replication is semi-conservative, meaning that each strand in the DNA acts as a template for the synthesis of a new complementary strand. Semi conservative because once DNA molecule is synthesized it has one strand from the parent and the other strand is a newly formed strand.
- DNA replication starts by taking one DNA molecule and giving two daughter molecules, with each newly synthesized molecule containing one new and one old strand.
- DNA replication simply is the process by which a DNA makes a copy of itself. Though easy as it may sound it’s a complex process happening inside of our cells, and many enzymes, proteins, and metal ions should work coherently to make this process happen.
Interesting Science Videos
Mechanism of DNA Replication
Summary: DNA replication takes place in three major steps.
- Opening of the double-stranded helical structure of DNA and separation of the strands
- Priming of the template strands
- Assembly of the newly formed DNA segments.
- During the separation of DNA, the two strands uncoil at a specific site known as the origin. With the involvement of several enzymes and proteins, they prepare (prime) the strands for duplication.
- At the end of the process, DNA polymerase enzyme starts to organize the assembly of the new DNA strands.
- These are the general steps of DNA replication for all cells but they may vary specifically, depending on the organism and cell type.
- Enzymes play a major role in DNA replication because they catalyze several important stages of the entire process.
- DNA replication is one of the most essential mechanisms of a cell’s function and therefore intensive research has been done to understand its processes.
- The mechanism of DNA replication is well understood in Escherichia coli, which is also similar to that in eukaryotic cells.
- In E.coli, DNA replication is initiated at the oriClocus (oriC), to which DnaA protein binds while hydrolyzing of ATP takes place.
Enzymes and Proteins Used in DNA Replication
Nucleases
- A nuclease is an enzyme that can cleave the phosphodiester bonds present in between the nucleotides.
- On the basis where they cleave, they are characterized as Exo and endonucleases.
- Exonucleases cleave nucleotides from their respective ends. Corresponding to this fact, these exonucleases show activity from both directions 5′ to 3′ and 3′ to 5′.
- Endonucleases act on the region in the middle of the targeted nucleotide. They are also endonucleases that are selective to which molecule they cleave and are sub-divided as DNase for DNA for cleaving and RNase for RNA cleaving. Additionally, recently discovered nucleases are also being used for gene editing such as Cas9 in the CRISPR genome editing technique.
- Restrictive endonuclease or restriction enzymes are the ones that cleave DNA into fragments at or near the specific recognition sites within the molecule known as restriction sites.
- To cleave the DNA, restriction endonuclease makes two incisions, once through each sugar-phosphate backbone of the DNA double helix. These endonucleases recognize a specific sequence of nucleotides and produce a double-stranded cut in the DNA.
- This specific sequence is usually 4 – 8 bases and is present in the recognition site.
DNA Polymerase
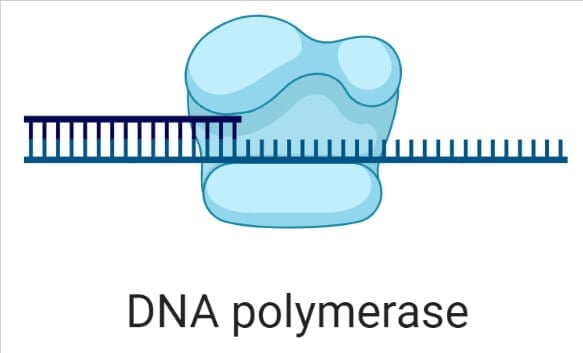
- DNA polymerases are the enzyme that is responsible for adding new nucleotides and synthesizing a new strand of DNA by taking the old fragmented strand as a template.
- DNA Polymerases also possess exonuclease activity, that cuts incorrectly added nucleotides, and allows the DNA replication to happen without errors.
- DNA Polymerase is of many types and functions based on the cell they are found in.
- In prokaryotic cells, there are three DNA polymerases: DNA Polymerase Ι, DNA Polymerase ΙΙ and DNA Polymerase ΙΙΙ.
- DNA polymerase Ι is a repair polymerase with 5′ to 3′ and 3′ to 5′ exonuclease activity. It is involved in the processing of Okazaki fragments during lagging strand synthesis.
- DNA polymerase ΙΙ has 3′ to 5′ exonuclease activity and participated in DNA repair with 5′ to 3′ polymerase activity.
- DNA polymerase ΙΙΙ is the primary enzyme involved in the DNA replication of E.coli. It has 3′ to 5′ exonuclease activity and 5′ to 3′ polymerase activity.
- In eukaryotic cells, there are five DNA polymerases: DNA Polymerase α, β, γ, δ and ε
- DNA polymerase α is a repair polymerase, with 3′ to 5′ exonucleases activities and 5′ to 3′ polymerase activities.
- DNA Polymerase β is a repair polymerase.
- DNA Polymerase γ shows polymerase activity 5′ to 3′ and exonucleases activity 3′ to 5′, it is involved in Mitochondrial DNA replication
- DNA Polymerase δ shows 3′ to 5′ exonuclease activity and 5′ to 3′ polymerase activity. This enzyme is involved in lagging strand synthesis.
- DNA Polymerase ε shows 3′ to 5′ and 5′ to 3′ exonucleases activities. This enzyme not only repairs but also synthesizes the leading strand efficiently in a 5′ to 3′ direction. It is the prime enzyme involved in DNA replication.
DNA ligase

- DNA ligase is a specific type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond.
- This enzyme joins the 3′ hydroxyl group of one nucleotide with the 5′ phosphate end of another nucleotide at an expense of ATP.
DNA helicase
- DNA helicase is a motor protein that moves directionally along a nucleic acid phosphodiester backbone, separating two nucleotides of DNA molecule.
- They separate double-stranded DNA molecules into single strands allowing each strand to be copied.
- During DNA replication, this DNA helicase unwinds DNA at the origin, a site where the replication is to be initiated.
- DNA helicase continues to unwind the double helix of DNA and thus forms a structure called replication fork, named after the forked appearance of two strands of DNA when unzipped apart.
- It is an energy-driven process as it involves the breaking of Hydrogen bonds between annealed nucleotide bases.
DNA primase
- Primase is an enzyme that is capable to synthesize short stretches of RNA sequences known as a primer.
- Primers are an integral part of DNA replication. These primers serve as an initiating site for the addition of nucleotides by DNA polymerase.
- DNA polymerase can only add nucleotide at pre-existing 3′ Hydroxyl group which is thus provided by the primers.
- As we can see that primers are short stretches of RNA, but replication is of DNA, so therefore after elongation of the chains of nucleotides, these primers are replaced by DNA.
DNA topoisomerase
- DNA topoisomerase is a class of enzymes that release helical tension during transcription and replication by creating transient nicks within the phosphate backbone on one or both strands of the DNA.
- This tension is aroused when the DNA molecule unwinds due to helicase activity and forms a replication fork. The progress of the replication fork generates supercoils, making it hard for other machinery involved to access the DNA molecule.
- Class Ι DNA topoisomerase makes a single-stranded break to relax the helix and progress the process.
- Class ΙΙ DNA topoisomerase break both the strands of DNA helix, this class of topoisomerases is also very important during the cell cycle for the condensation of chromosomes.
Single strand binding proteins
- The single-strand binding (SSB) protein are DNA binding proteins, that binds to single-stranded DNA to facilitate DNA replication.
- SSB proteins prevent the hardening of strands during DNA replication. It also protects strands from nuclease degradation and prevents the rewinding of DNA.
- These proteins destabilize helical duplexes so that DNA polymerase can hold onto the DNA during DNA replication, recombination, and repair.
- It also removes unwanted secondary structures on strands for easy access of the strands to the machinery involved in DNA replication.
- Thus, SSB proteins stabilize the single-stranded DNA structure that is important for genomic progression.
So, in summary here are the list of 7 enzymes and proteins used in DNA Replication:
- Nucleases
- DNA Polymerase
- DNA ligase
- DNA helicase
- DNA primase
- DNA topoisomerase
- Single strand binding proteins
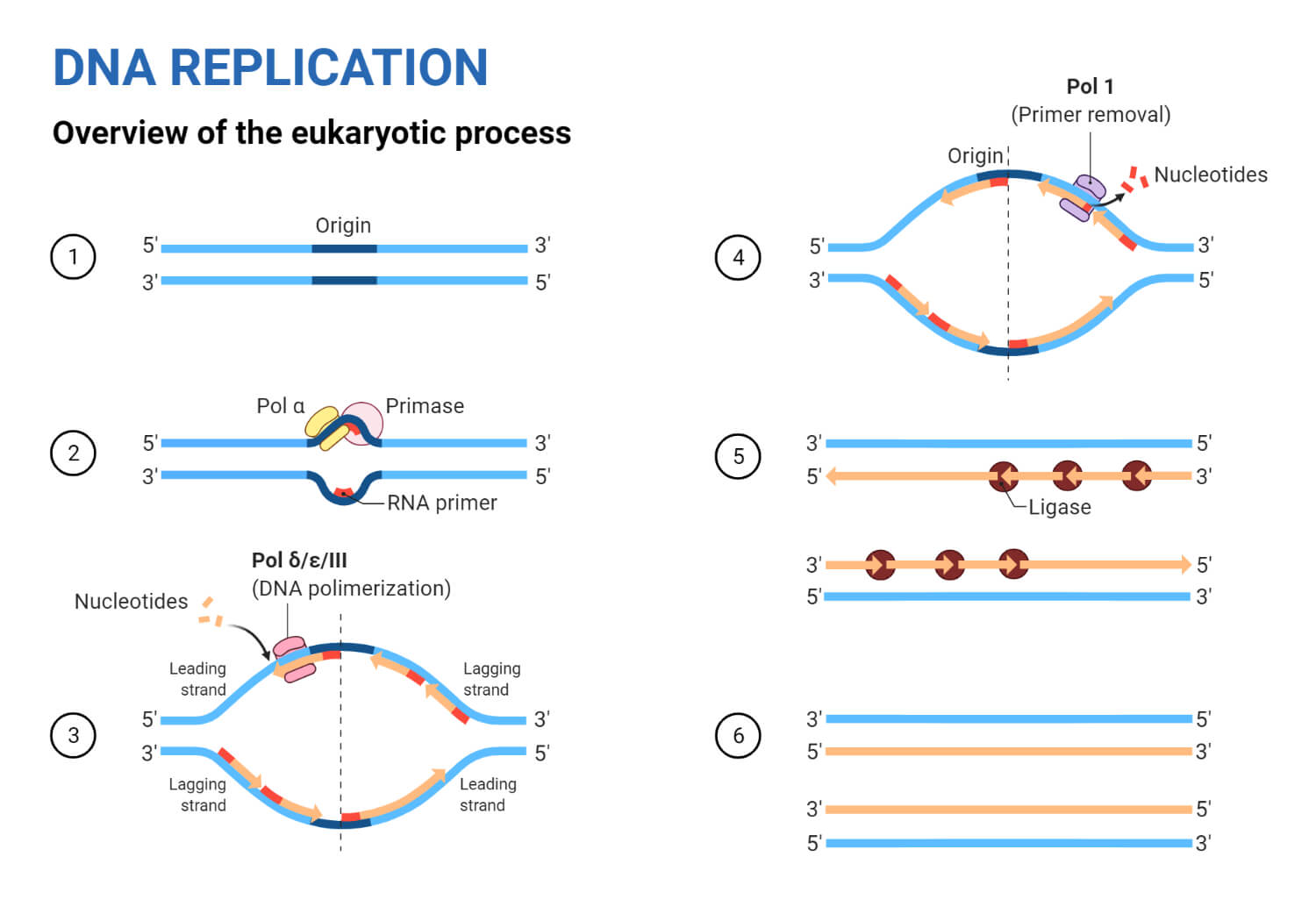
Steps in DNA Replication
Step 1: Formation of Replication Fork
- Before DNA can replicate, this double-stranded molecule must unwind into two single strands to initiate the replication process.
- DNA unwinds when the complementary base pairing between the double-stranded is broken, and the site to initiate this unwinding is denoted by specific regions (Adenine and Thymine rich).
- These specific coding regions are referred to as Origin of Replication (Ori) and thus the replication process begins.
- These origins are targeted by initiator proteins, which go on to recruit more proteins that can help the replication process by forming a replication fork around the Ori.
- Within this replication protein complex is an enzyme DNA helicase, which starts to unwind the DNA from its Ori and exposes two strands resembling a Y-like structure referred to as replication fork.
- The activity of helicase causes topological stress to the un-winded strand forming supercoiled DNA, this stress is relieved by Topoisomerase by negative supercoiling.
- The replication fork is bidirectional; one strand is oriented to 3′ to 5′ direction (leading strand) and the other strand is oriented to 5′ to 3′ direction (lagging strand) but the addition of nucleotide progress only in 5′ to 3′ direction.
- The formation of a replication fork exposing two single-stranded strands marks the beginning of Initiation.
Step 2: Initiation
- One strand runs from 3′ to 5′ direction towards the replication fork and is referred to as leading strand and the other strand runs from 5′ to 3′ away from the replication fork and is referred to as lagging strands.
- To this exposed single-stranded DNA, SSB proteins are adhered to prevent recoiling of DNA and to stabilize it.
- After which another enzyme DNA primase comes into action to synthesize a short stretch of RNA primer, which provides a free 3′ hydroxyl group for DNA polymerase can now add nucleotides and extend the new chain of nucleotides.
Step 3: Elongation
- Now that primer is added to unzipped two single-stranded DNA, these strands now act as a template for synthesizing new DNAs.
- The enzyme DNA polymerase synthesizes new nucleotide to match the template and add on to the free 3′ hydroxyl group provided by the primer in each single-stranded DNA.
- The leading strand runs from 3′ to 5′ so the addition of nucleotides by DNA polymerase happens from 5′ to 3′ direction. As the replication fork progresses the addition of nucleotide is continuous thus only requiring the primer once.
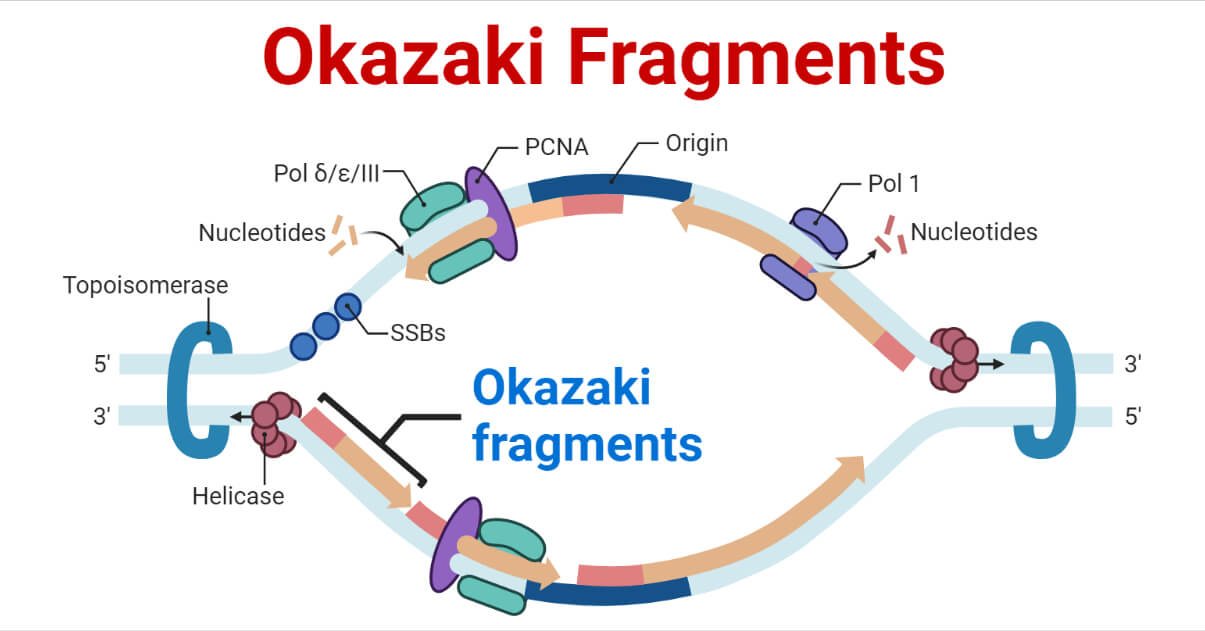
- However, lagging strands is antiparallel and run from the 5′ to 3′ direction, the continuous addition of nucleotides is not possible as the replication fork progresses, DNA polymerase cannot add complementary nucleotides to the 5′ end. Therefore, multiple primers are required.
- Due to this phenomenon, the DNA nucleotides synthesis from lagging strands occurs in fragments. These fragments are termed Okazaki fragments.
- Hence, the leading strand using only one primer synthesizes nucleotides continuously, while the lagging strand uses multiple primers and thus synthesizes nucleotides discontinuously.
Step 4: Termination
- RNA primers of both leading and lagging strands are cleaved out or degraded by exonucleases activity of DNA polymerase, and the nicks or gaps so formed are filled with DNA and sealed by the enzyme DNA ligase.
- DNA polymerase also shows proofreading activity and check, remove and replace any errors.
- Interestingly, in eukaryotic organisms having linear DNA, when RNA primer at 5′ end of daughter strand is removed, there is not a preceding 3′ OH such that DNA polymerase can use it to replace with DNA.
- So, at the 5′ end of daughter strands, there is a gap (missing DNA). This missing DNA can cause a loss of information contained in that region. This gap must be filled before the next round of replication.
- For solving this end replication problem, researchers have found that linear ends of DNA called telomere are used which contain specific G: C rich repeats. These sequences are known as telomere sequences.
- These telomere sequences do not code anything but are essential to fill in the gap in the daughter strand and maintain the integrity of DNA.
- Eventually, the replication forks terminate at terminating recognizing sequences (ter).
- The ter sequences are of around 23 base pairs which facilitate as the binding sites for TUS protein.
- This ter- TUS complex arrest replication fork and terminate.
So, in summary, these are the 4 steps of DNA Replication:
- Formation of Replication Fork
- Initiation
- Elongation
- Termination
Okazaki Fragments
- The two DNA strands run in opposite or antiparallel directions, and therefore to continuously synthesize the two new strands at the replication fork requires that one strand is synthesized in the 5’to3′ direction while the other is synthesized in the opposite direction, 3’to 5′.
- However, DNA polymerase can only catalyze the polymerization of the dNTPs only in the 5’to 3’direction.
- This means that the other opposite new strand is synthesized differently. But how?
- By the joining of discontinuous small pieces of DNA that are synthesized backward from the direction of movements of the replication fork. These small pieces or fragments of the new DNA strand are known as the Okasaki Fragments.
- The Okasaki fragments are then joined by the action of DNA ligase, which forms an intact new DNA strand known as the lagging strand.
- The lagging phase is not synthesized by the primer that initiates the synthesis of the leading strand.
- Instead, a short fragment of RNA serves as a primer (RNA primer) for the initiation of replication of the lagging strand.
- RNA primers are formed during the synthesis of RNA which is initiated de novo, and an enzyme known as primase synthesizes these short fragments of RNA, which are 3-10 nucleotides long and complementary to the lagging strand template at the replication fork.
- The Okazaki fragments are then synthesized by the extension of the RNA primers by DNA polymerase.
- However, the newly synthesized lagging strand is that it contains an RNA-DNA joint, defining the critical role of RNA in DNA replication.
Replication Fork Formation and its function
- The replication fork is the site of active DNA synthesis, where the DNA helix unwinds and single strands of the DNA replicates.
- Several sites of origin represent the replication forks.
- The replication fork is formed during DNA strand unwinding by the helicase enzyme which exposes the origin of replication. A short RNA primer is synthesized by primase and elongation done by DNA polymerase.
- The replication fork moves in the direction of the new strand synthesis. The new DNA strands are synthesized in two orientations, i.e 3′ to 5′ direction which is the leading strand, and the 5′ to 3′ orientation which is the lagging strand.
- The two sides of the new DNA strand (leading and lagging strand) are replicated in two opposite directions from the replication fork.
- Therefore the replication fork is bi-directional.
Leading Strand
- The leading strand is the new DNA strand that is continuously synthesized by the DNA polymerase enzyme.
- It is the simplest strand that is synthesized during replication.
- The synthesis starts after the DNA strand has unzipped and separated. This generates a short piece of RNA known as a primer, by the DNA primase enzyme.
- The primer binds to the 3′ end (start) of the strand, thus initiating the synthesize of the new strand (leading strand).
- The synthesis of the leading strand is a continuous process.
The Lagging Strand
- This is the template strand (5′ to 3′) that is synthesized in a discontinuous manner by RNA primers.
- During the synthesis of the leading strand, it exposes small, short strands, or templates that are then used for the synthesis of the Okasaki fragments.
- The Okasaki fragments synthesize the lagging strand by the activity of DNA polymerase which adds the pieces of DNA (the Okasaki fragments) to the strand between the primers.
- The formation of the lagging strand is a discontinuous process because the newly formed strand (lagging strand) is the fragmentation of short DNA strands.
Applications of DNA Replication
- DNA replication makes the transfer of genetic information from one generation to another possible.
- It is an important phenomenon happening inside our cells, that allows the body to maintain homeostasis and integrity of the body.
- With the available information about DNA replication, scientific communities today have a proper idea of genome sequencing which has now been applied in different expertise ranging from clinical diagnosis to possible treatment of genetic diseases.
- DNA replication has made sequencing of whole human genome sequencing possible.
- Cloning of genes has also been possible by DNA replication.
- Enzymes involved in DNA replication have now been greatly studied due to their wider applications. The recent breakthrough Cas9/ CRISPR technology where nucleases are used to cleave the desired portion of DNA and replace it with required nucleotides is the prime example of how we can use these enzymes and make potential advancements in them thus broadening and exploring their uses.
- Polymerase Chain Reaction uses DNA polymerases to repeatedly replicate DNA in-vitro and has numerous applications in diagnosis, sequencing, and recombinant DNA technology.
- The formation of complementary DNA (cDNA) can also be considered as an example of a wider application of the enzymes involved in DNA replication.
- There are various applications of DNA replication, we can even consider that if there is any technique involving genes, some way or the other DNA replication is applied.
DNA Replication Stress
During DNA replication, the process and the DNA genome undergoes various stress arising from the mechanism. these stresses an result in stalled replication and stalled replication fork formation. Several events contribute to these stresses, including;
- Unusual DNA structure
- Mismatched ribonucleotides
- Tensions arising from concurrent mechanisms of replication and transcription
- Inadequate availability of important replication factors
- Fragile sites on the replicating DNA strand
- Overexpression or constitutive activation of oncogenes
- Inaccessible chromatins
Kinase regulatory proteins such as ATM (ATM serine/threonine kinase) and ATP are proteins that assist in alleviating replication stress. These proteins get recruited and activated by DNA damages.
Stalled replication forks may collapse if the regulatory proteins do not stabilize, and if and when this happens, initiation of repairing mechanisms to reassembling of the replication fork takes place. this helps to amend damages the damaged ends of DNA.
Similarities between Prokaryotic and Eukaryotic DNA Replication
- The unwinding mechanism of DNA before replication is initiated is the same for both Prokaryotes and eukaryotes.
- In both organisms, the DNA polymerase enzyme coordinated the synthesis of new DNA strands.
- Additionally, both organisms use the semi-conservative replication pattern, making the leading and lagging strands in different directions. Okasaki fragments make the lagging strand.
- Lastly, both organisms initiate DNA replication using a short RNA primer.
Eukaryotic vs. Prokaryotic DNA Replication (11 Major Differences)
| S.N. | Eukaryotic DNA Replication | Prokaryotic DNA replication |
| 1. | Occurs in eukaryotic cells. | Occurs in a prokaryotic cell. |
| 2. | This process takes place in the cell’s nucleus. | This process takes place in the cell’s cytoplasm. |
| 3. | There are multiple sites for the origin of replication per DNA molecule. | There is a single site for the origin of replication per DNA molecule. |
| 4. | Initiation of DNA replication is carried out by multi-subunit proteins, origin recognition complex. | Initiation of DNA replication is carried out by protein DnaA and DnaB. |
| 5. | Multiple replication forks are formed in a DNA molecule. | Only two replication forks are formed in a DNA molecule. |
| 6. | Okazaki fragments are short of around 100-200 nucleotides in length | Okazaki fragments are large, around 1000-2000 nucleotides in length. |
| 7. | It is a slow process with around 100 nucleotides added per second. | It is a fast process with around 2000 nucleotides added per second. |
| 8. | DNA is linear and double-stranded. | DNA is circular and double-stranded. |
| 9. | DNA polymerase involved in eukaryotic DNA replication is DNA polymerases ε, α, and δ. | DNA polymerase involved in prokaryotic DNA replication is DNA polymerase Ι, and ΙΙΙ. |
| 10. | Eukaryotic cells have telomeres at the end of DNA thus they are replicated. | Prokaryotic cells have circular DNA thus they are not replicated. |
| 11. | DNA gyrase (Telomerase) is needed. | DNA gyrase (Telomerase) is not needed. |
References
- Weiguo Cao, “DNA Ligases: Structure, Function, and Mechanism”, Current Organic Chemistry 2002; 6(9). https://doi.org/10.2174/1385272023373950
- https://www.nature.com/scitable/definition/helicase-307/
- https://proteinswebteam.github.io/interpro-blog/potm/2006_1/Page2.htm
- https://www.yourgenome.org/facts/what-is-dna-replication
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/replication/a/molecular-mechanism-of-dna-replication
- https://teachmephysiology.com/biochemistry/cell-growth-death/dna-replication/
- https://courses.lumenlearning.com/wm-biology1/chapter/reading-major-enzymes/
- https://geneticeducation.co.in/single-stranded-binding-protein-ssb-structure-and-function/
- https://www.thoughtco.com/dna-replication-3981005
- https://byjus.com/biology/difference-between-prokaryotic-and-eukaryotic-replication/
- https://www.vedantu.com/biology/difference-between-prokaryotic-and-eukaryotic-dna-replication
- https://www.majordifferences.com/2013/03/difference-between-prokaryotic-and.html
