DNA microarrays are solid supports, usually of glass or silicon, upon which DNA is attached in an organized, pre-determined grid fashion.
Each spot of DNA, called a probe, represents a single gene.
DNA microarrays can analyze the expression of tens of thousands of genes simultaneously.
There are several synonyms for DNA microarrays, such as DNA chips, gene chips, DNA arrays, gene arrays, and biochips.
Interesting Science Videos
Principle of DNA Microarray Technique
- The principle of DNA microarrays lies on the hybridization between the nucleic acid strands.
- The property of complementary nucleic acid sequences is to specifically pair with each other by forming hydrogen bonds between complementary nucleotide base pairs.
- For this, samples are labeled using fluorescent dyes.
- At least two samples are hybridized to chip.
- Complementary nucleic acid sequences between the sample and the probe attached on the chip get paired via hydrogen bonds.
- The non-specific bonding sequences while remain unattached and washed out during the washing step of the process.
- Fluorescently labeled target sequences that bind to a probe sequence generate a signal.
- The signal depends on the hybridization conditions (ex: temperature), washing after hybridization etc while the total strength of the signal, depends upon the amount of target sample present.
- Using this technology the presence of one genomic or cDNA sequence in 1,00,000 or more sequences can be screened in a single hybridization.
Types of DNA Microarrays
There are 2 types of DNA Chips/Microarrays:
- cDNA based microarray
- Oligonucleotide based microarray
Spotted DNA arrays (“cDNA arrays”)
- Chips are prepared by using cDNA.
- Called cDNA chips or cDNA microarray or probe DNA.
- The cDNAs are amplified by using PCR.
- Then these immobilized on a solid support made up of nylon filtre of glass slide (1 x 3 inches). The probe DNA are loaded into a a spotting spin by capillary action.
- Small volume of this DNA preparation is spotted on solid surface making physical contact between these two.
- DNA is delivered mechanically or in a robotic manner.
Oligonucleotide arrays (Gene Chips)
- In oligonucleotide microarrays, short DNA oligonucleotides are spotted onto the array.
- Small number of 20-25mers/gene.
- The main feature of oligonucleotide microarray is that each gene is normally represented by more than one probe.
- Enabled by photolithography from the computer industry
- Off the shelf
Requirements of DNA Microarray Technique
There are certain requirements for designing a DNA microarray system, viz:
- DNA Chip
- Target sample (Fluorescently labelled)
- Fluorescent dyes
- Probes
- Scanner
Steps Involved in cDNA based Microarray
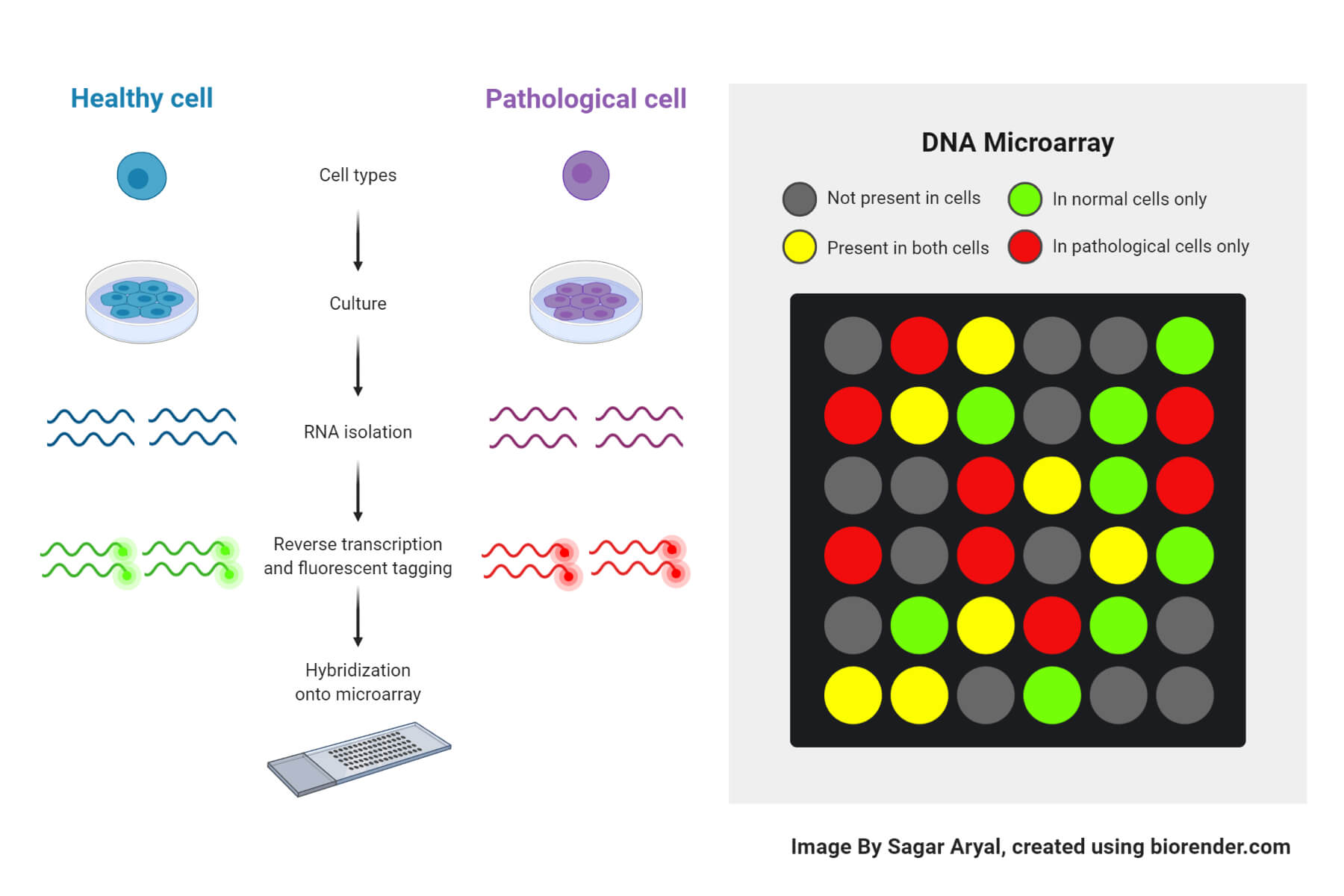
The reaction procedure of DNA microarray takes places in several steps:
- Collection of samples
- The sample may be a cell/tissue of the organism that we wish to conduct the study on.
- Two types of samples are collected: healthy cells and infected cells, for comparison and to obtain the results.
- Isolation of mRNA
- RNA is extracted from the sample using a column or solvent like phenol-chloroform.
- From the extracted RNA, mRNA is separated leaving behind rRNA and tRNA.
- As mRNA has a poly-A tail, column beads with poly-T-tails are used to bind mRNA.
- After the extraction, the column is rinsed with buffer to isolate mRNA from the beads.
- Creation of labeled cDNA
- To create cDNA (complementary DNA strand), reverse transcription of the mRNA is done.
- Both the samples are then incorporated with different fluorescent dyes for producing fluorescent cDNA strands. This helps in distinguishing the sample category of the cDNAs.
- Hybridization
- The labeled cDNAs from both the samples are placed in the DNA microarray so that each cDNA gets hybridized to its complementary strand; they are also thoroughly washed to remove unbounded sequences.
- Collection and analysis
- The collection of data is done by using a microarray scanner.
- This scanner consists of a laser, a computer, and a camera. The laser excites fluorescence of the cDNA, generating signals.
- When the laser scans the array, the camera records the images produced.
- Then the computer stores the data and provides the results immediately. The data thus produced are then analyzed.
- The difference in the intensity of the colors for each spot determines the character of the gene in that particular spot.
Applications of DNA Microarray
In humans, they can be used to determine how particular diseases affect the pattern of gene expression (the expression profile) in various tissues, or the identity (from the expression profile) of the infecting organism. Thus, in clinical medicine alone, DNA microarrays have huge potential for diagnosis.
Besides, it has applications in many fields such as:
- Discovery of drugs
- Diagnostics and genetic engineering
- Alternative splicing detection
- Proteomics
- Functional genomics
- DNA sequencing
- Gene expression profiling
- Toxicological research (Toxicogenomics)
Advantages of DNA Microarray
- Provides data for thousands of genes in real time.
- Single experiment generates many results easily.
- Fast and easy to obtain results.
- Promising for discovering cures to diseases and cancer.
- Different parts of DNA can be used to study gene expression.
Disadvantages of DNA Microarray
- Expensive to create.
- The production of too many results at a time requires long time for analysis, which is quite complex in nature.
- The DNA chips do not have very long shelf life.
References
- https://www.slideshare.net/Haddies/dna-microarray-dna-chips
- https://www.slideshare.net/mahendrareddy59/dna-microarray-56074935
- https://www.news-medical.net/life-sciences/DNA-microarray.aspx
- https://www.researchgate.net/figure/Steps-in-DNA-Microarray-Experiment-III-GENE-EXPRESSION-DATA-A-typical-DNA-microarray_fig1_236868392
- https://bitesizebio.com/7206/introduction-to-dna-microarrays/
- http://www.highveld.com/molecular-biology/oligonucleotide-microarrays.html
- https://courses.cs.washington.edu/courses/cse527/04au/slides/lect02.pdf
