Prokaryotic topoisomerase II, also known as DNA gyrase, is the only topoisomerase that introduces negative supercoils into DNA thus far.
DNA topoisomerases are a class of enzymes that can change the topological state of closed-circular DNA molecules. There are two classes of topoisomerase enzymes. They are:
- Type I
- Type II
DNA topoisomerases have been isolated from viral, prokaryotic, and eukaryotic sources.
The thermophilic bacterium Sulfolobus produced DNA gyrase, a “reverse gyrase” that adds positive superhelical twists to DNA in the presence of ATP. Since it is lacking in higher eukaryotes yet necessary for all bacteria, it is a desirable target for antibiotics.
Discovery of DNA Gyrase
When seeking to identify the Escherichia coli host components necessary for bacteriophage λ site-specific integration, Gellert and co-workers made the initial discovery of DNA gyrase in 1976.
The work on two types of DNA synthesis inhibitors, the quinolones (nalidixic acid, oxolinic acid, and ciprofloxacin) and the coumarins (novobiocin, coumermycin, and chlorobiocin), had been done before the discovery of gyrase. Since then, DNA gyrase has been the target of these antibacterial classes.
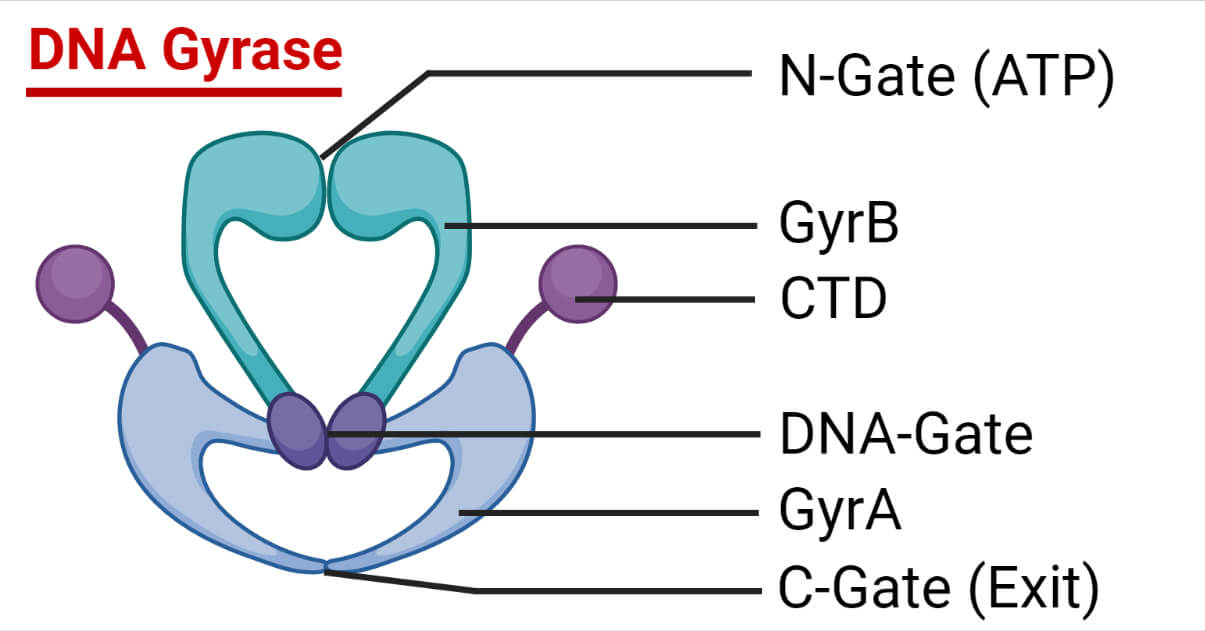
Structure of DNA Gyrase
DNA Gyrase is made up of two distinct proteins that are each coded for by genes that have previously been found to be genetic loci determining either nalidixic acid or coumermycin resistance (nalA and cou).
With the discovery of gyrase, these genes are now referred to as gyrA and gyrB (molecular mass ≈96 kDa and 88 kDa, respectively) and are arranged as an A2B2 tetramer.
The tyrosine utilized in DNA cleavage and ligation is found in gyrA.
The ATP binding site is found in gyrB.
Reactions of DNA Gyrase
Numerous topological interconversions of DNA molecules are carried out by DNA gyrase. In addition to the negative supercoiling reaction, gyrase may catenate and decatenate two duplex DNA circles and relax negatively supercoiled DNA without ATP.
Supercoiling and Relaxation
The DNA supercoiling reaction is triggered by spermidine and requires ATP in addition to a divalent cation like Mg2+. The enzyme DNA gyrase can spirally wind up DNA. At 30°C, one molecule has been estimated to cause a linking number difference of roughly 100/min.
Negatively supercoiled DNA will be relaxed by gyrase in the absence of ATP. The supercoiling process is substantially more efficient than the relaxation activity of gyrase, requiring 20 to 40 times as much enzyme to achieve a similar rate.
Catenation, Decatenation, and Unknotting
Gyrase can unknot DNA that has been knotted and can catalyze the production and resolution of DNA catenanes. Gyrase should theoretically be able to knot rings of double-stranded DNA as well. However, this reaction has not been observed. The drugs coumarin and quinolone block these ATP-dependent processes.
Spermidine stimulates catenation and decatenation. Under the right DNA concentration regimes, catenation and decatenation processes are projected not to need ATP hydrolysis since they are energetically advantageous. However, it is observed that ATP is needed for both of these processes.
Models for the Gyrase-DNA Complex
Kirchhausen and Krueger have proposed the most intricate models of the gyrase-DNA complex based on their interpretations of electron microscopy and scattering data. Kirchhausen suggested that the gyrase complex is structured like a heart, with the DNA wrapping around the protein so that its center is situated in the space between the top lobes of the heart.
About 120 bp of DNA are wrapped around the protein in the gyrase-DNA complex. The DNA tails are thought to be at an angle of 120⁰ and are placed near the DNA entry and exit points. The gyrase particle has been estimated to measure 175 Å by 52 Å in size.
Mechanism of DNA Supercoiling by DNA Gyrase
All topoisomerase reactions require the binding of the protein to DNA, DNA cleavage, strand passage, DNA reunion, and in some cases, ATP hydrolysis, and the enzymes are likely to have a similar mode of action to gyrase. Despite adhering to the general topoisomerase mechanism, DNA gyrase also needs special mechanistic characteristics that govern its capacity to supercoil DNA actively.
The observed reactions of DNA gyrase are listed below:
- ATP-dependent negative supercoiling of closed-circular double-stranded DNA
- ATP-independent relaxation of negatively supercoiled DNA
- Nucleotide-dependent relaxation of positively supercoiled DNA
- Formation and resolution of catenated DNA
- Resolution of knotted DNA
- Quinolone or calcium ion-induced double-stranded breakage of DNA
- DNA-dependent ATP hydrolysis
Each of the above reactions is a part of a single mechanism that can occur with various substrates or under various circumstances.
Gyrase interaction with Antibiotics
Gyrase is an excellent therapeutic target due to its importance in bacterial cells and the apparent absence of gyrase activity in eukaryotes.
Most of these can be classified into two groups, quinolones and coumarins. Nevertheless, other compounds don’t fit into either of these categories.
Quinolone Drugs
Numerous techniques have taken advantage of quinolones’ capacity to cause DNA breakage by gyrase. The gyrase-induced DNA cleavage reaction can evaluate the effectiveness of a certain drug as a gyrase inhibitor. This reaction has been interpreted as proof that a double-stranded cleavage event occurred as part of the regular mechanism for DNA supercoiling. It is hypothesized that quinolone drugs prevent the DNA cleavage phase of the supercoiling process.
Coumarin drugs
The DNA gyrase enzyme was identified as the coumarin drugs’ primary target. The precise mechanism by which coumarins inhibit the gyrase ATPase process is not established. As per steady-state kinetic experiments, novobiocin and coumermycin are likely to be competitive inhibitors of ATP hydrolysis by gyrase and DNA supercoiling.
Other Drugs
Outside of the quinolone and coumarin families, very few gyrase inhibitors exist. Recent research has revealed that two antibiotics different from quinolones and coumarins may target DNA gyrase inside cells.
Cinodine is an antibiotic of the glycocinnamoylspermidine class made by the Nocardia species. In vitro experiments with Micrococcus luteus DNA gyrase has demonstrated that this drug binds to DNA and prevents DNA supercoiling.
Microcin B17 is effective against a wide variety of enterobacteria. It has been demonstrated to be a DNA replication inhibitor, which causes a quick arrest in DNA synthesis, the triggering of the SOS response, DNA breakdown, and cell death.
In Vivo Role of Gyrase
Gyrase was first identified as a host factor necessary for the bacteriophage’s λ site-specific integration. Still, it has now been linked to transcription, DNA replication, modulation of chromosomal superhelical tension, and a plethora of other cellular functions.
DNA Replication
For DNA replication to occur in vivo, DNA gyrase is essential. Given the DNA unwinding at the replication fork, it would be expected that negative supercoiling would facilitate replication.
The same antibiotics that inhibit DNA replication also inhibit gyrase. Additionally, temperature-sensitive mutations of the gyrA and gyrB genes in E. coli prevent DNA replication when the temperature is too high.
gyrA and gyrB mutations appear to have differing effects on several replication processes. While a gyrA mutant caused a swift stop of chain elongation, gyrB mutant was found to inhibit the initiation of replication but not chain elongation.
Transcription
Drugs that decrease DNA gyrase activity can impact the degree of gene expression. It has been found that negatively supercoiled DNA has improved transcriptional capacity compared to relaxed, nicked, or linear DNA.
Negative supercoiling can both activate and inhibit transcription. Therefore, it has been proposed that gyrase’s function during transcription is to relax the positive supercoils before the transcription complex. In contrast, topoisomerase I relax the negative supercoils in the back.
References
- Collin, F., Karkare, S., & Maxwell, A. (2011). Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Applied microbiology and biotechnology, 92(3), 479–497. https://doi.org/10.1007/s00253-011-3557-z
- Reece, R. J., & Maxwell, A. (1991). DNA gyrase: structure and function. Critical reviews in biochemistry and molecular biology, 26(3-4), 335–375. https://doi.org/10.3109/10409239109114072
- Renier Vélez-Cruz and Neil Osheroff. (2004). DNA Topoisomerases: Type II. In Encyclopedia of Biological Chemistry, pg. 806-811. Elsevier Inc.
