The local alterations in chromatin structure facilitate the assembly of the transcription factors and the RNA polymerase holoenzyme at the site of the promoter. The two most important ways for altering chromatin structures are covalent histone modifications and nucleosome remodeling.
What is Covalent Histone Modification?
Covalent Histone Modification is associated with the structural changes that occur at the time of replication and transcription. These modifications are mediated by chromatin modification complexes, which are multiprotein complexes that modify histones post-translationally. All of the histone proteins are modified chemically. The N- terminal tails of the histones are subjected to a wide range of post-translational covalent modifications. These modifications include Acetylation, histone tails undergoing methylation, Sumoylation, Ubiquitylation, ADP ribosylation, Glycosylation, and Biotinylating. The majority of these modifications occur in the tail regions, but there are occasional modifications within the histone fold( methylation of lysine at the 79th position of histone H3).
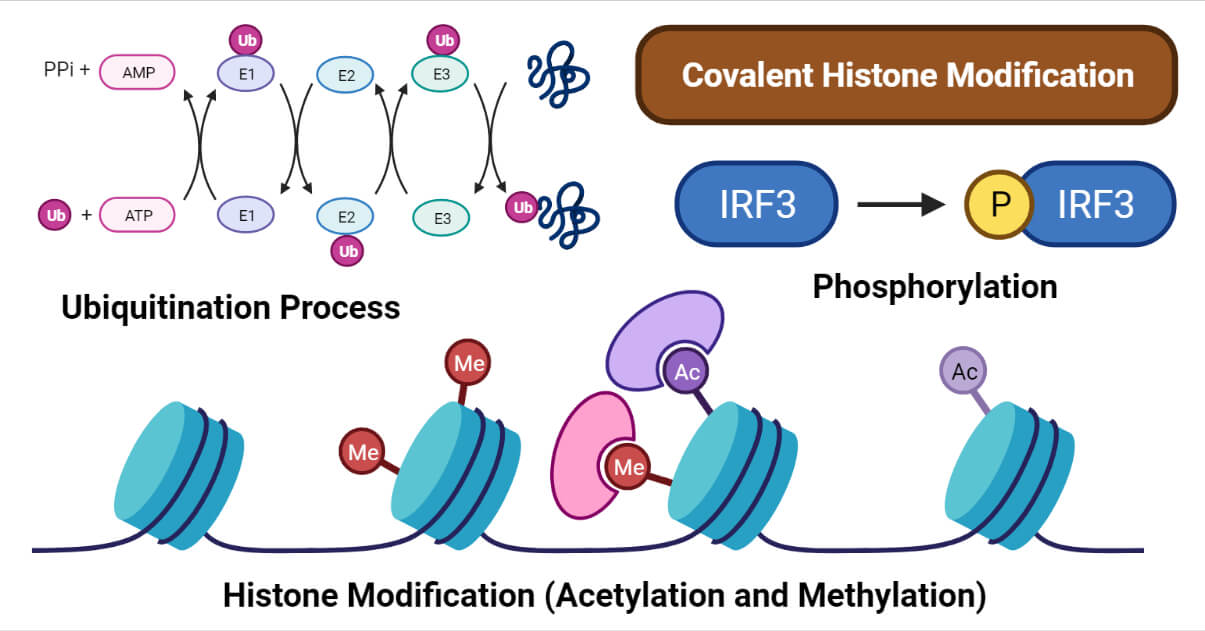
The most common types of post-translational chemical modifications of histone are described below.
Acetylation
Each of the histone proteins making up the nucleosome core contains a flexible N- terminus of 11-37 amino acid residues extending from the fixed structure of the nucleosome i.e N- termini are known as histone tails (Hayes JJ 2001).
Acetyl groups are added to the lysine amino acid residues in the histone tail of each of the core molecules. The enzyme associated with it is called histone acetyltransferases(HATs) also known as histone acetylases. A number of different histone acetyltransferases have been identified and are distinguished by their abilities to target different histones. When a histone is acetylated, an acetyl group is added to one or more of its lysine residues at the epsilon amino group, by removing the positive charge. It reduces the affinity of the histone for DNA and possibly also reduces the interaction between the individual nucleosome that leads to the formation of the 30nm chromatin fiber. The process of acetylation is reversible.
The enzyme histone deacetylases (HDACs) catalyze the removal of acetyl groups(deacetylation)(Seto E, 2014). The histones in heterochromatin are generally deacetylated whereas the euchromatin is acetylated. Acetylation of histones also creates a specific binding site for binding sites for other proteins.
However, the proteins that bind to lysine contain a domain called BROMODOMIAN. However, it is a ubiquitous and important post-translation; the modification that regulates protein structure and function (Drazic et al., 2016). While most of the cellular protein acylation is facilitated and mediated by acetyltransferases, non-enzymatic induced by activated esters or anhydrides are widely reported (Wagner and Hirschey, 2014).
Common deacetylases such as SIRT2 and SIRT3 (Wagner and Hirschey, 2014). These fall in the category of cofactor NAD+ – dependent, the metabolic disorder of NAD+ and NADH biosynthesis will also influence the process of deacetylation regulation of cells ( Wagner and Hirschey, 2014 ; Drazic et al, 2016)
Methylation
Most of the lysine and arginine at the N- terminal histone tails participates methylation. These may vary such as mono- di- or trimethyl for lysine and mono- or dimethyl for arginine’s. Unlike acetylation and phosphorylation, however histone methylation does not alter the charge of the histone protein (Bannister, A., 2011).
Histone methylation is catalyzed by histone methyltransferases(HMTs) and histone demethylases(HDMs) remove these modifications. The different parts of the N- terminal tails of H3 and H4 histone are associated with both activation and repression, depending on the particular amino acid that is modified in the histone tail. For example, methylation of lysine at the 9th of H3(H3K9) is also associated with gene silencing and condensation of chromatin whereas methylation of lysine at 4th position of H3(H3K4) is associated with transcription activation (Zhang Xiaoli, et al., 2016).
The methylation of histone provides a site for the binding of other sites leading to the alteration of chromatin structure. The proteins that bind to methylated histone contain a domain called the Chromo domain (called chromatin organization modifier)(Mariño-Ramírez, et al., 2005)Histone methylation plays a very fundamental role in heterochromatin formation, chromosome X- inactivation, genomic imprinting, and transcriptional regulation.
Phosphorylation
The concept of phosphorylation shows its significant impact on cellular activity. It takes place in serine, threonine, and tyrosine present in the N- terminal histone tails. The levels of modification are controlled by kinases and phosphatases which can add and remove the phosphate group. All of the identified histones transfer the hydroxyl group to phosphate(kinases) or remove it from it(phosphatases) which affects downstream activities. Likewise, phosphorylation of histone also serves as a recruiting point for DNA damage and repair proteins.
ADP ribosylation
The addition of ADP ribose can be detected in all the four core histones as well as linked histone H1. These modifications are known to level up to an increase in DNA damage implicating DNA damage response.
Ubiquitylation
It involves the ubiquitination of histone with a 76 amino acid polypeptide which is attached to histone lysine (Ubiquitination (Ubiquitylation) (news-medical.net))
Sumoylation
The addition of SUMO protein (Small Ubiquitin-related Modifier) is a polypeptide of 97 amino acid residues added to the carboxyl group and amino groups that target lysine in a target protein. Sumoylation has been detected on all four core histones and functions by antagonizing acetylation and ubiquitylation. It has been associated with repressive functions.
Functions
- These modifications occur in chromatin remodeling.
- The various process like synapsis, double-stranded break formation, and repair undergoes covalent modifications.
- It allows participation in transcription and recombination.
References
- Bannister, A., Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res 21, 381–395 (2011). https://doi.org/10.1038/cr.2011.22for example
- Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864:1372–1401.
- Johnson, Sean, and Shin-Ichiro Imai. “NAD + biosynthesis, aging, and disease.” F1000Research vol. 7 132. 1 Feb. 2018, doi:10.12688/f1000research.12120.1
- Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev. 2001 Apr;11(2):124-9. doi: 10.1016/s0959-437x(00)00168-4. PMID: 11250133.
- Mariño-Ramírez, Leonardo et al. “Histone structure and nucleosome stability.” Expert review of proteomics vol. 2,5 (2005): 719-29. doi:10.1586/14789450.2.5.719
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi: 10.1101/cshperspect.a018713. PMID: 24691964; PMCID: PMC3970420.
- Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16.3 Ubiquitination (Ubiquitylation) (news-medical.net)
- Zhang Xiaoli, Liu Xinqiang, Zhao Yanli, Cheng Jiasen, Xie Jiatao, Fu Yanping, Jiang Daohong, Chen Tao, Histone H3 Lysine 9 Methyltransferase DIM5 Is Required for the Development and Virulence of Botrytis cinerea, Frontiers in Microbiology , 7 , 2016 https://www.frontiersin.org/article/10.3389/fmicb.2016.01289, 10.3389/fmicb.2016.01289 ISSN=1664-302X
