- The genetic code is the set of rules by which a linear sequence of nucleotides specifies the linear sequence of a polypeptide.
- That is, they specify how the nucleotide sequence of an mRNA is translated into the amino acid sequence of a polypeptide.
- Thus, the relationship between the nucleotide sequence of the mRNA and the amino acid sequence of the polypeptide is the genetic code.
- The nucleotide sequence is read as triplets called codons.
PRINCIPLES OF THE GENETIC CODE
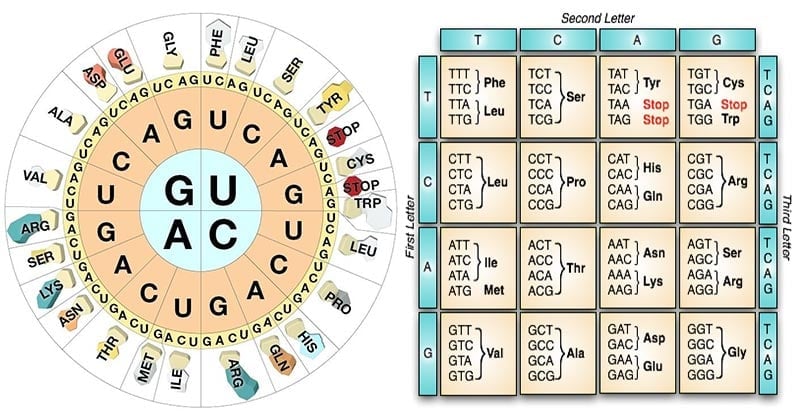
- The genetic code consists of 64 different codons, each of which codes for 1 of the 20 amino acids.
- A codon consists of a triplet of nucleotide bases.
Interesting Science Videos
CHARACTERISTICS OF THE GENETIC CODE
The genetic code is endowed with many characteristic properties which have actually been proved by definite experimental evidences.
- Triplet nature:
- Singlet and doublet codes are not adequate to code for 20 amino acids; therefore, it was pointed out that triplet code is the minimum required.
- Degeneracy
- The code is degenerate which means that the same amino acid is coded by more than one base triplet.
- Degeneracy does not imply lack of specificity in protein synthesis.
- It merely means that a particular amino acid can be directed to its place in the peptide chain by more than one base triplets.
- For example, the three amino acids arginine, alanine and leucine each have six synonymous codons.
- The code degeneracy is basically of 2 types: partial and complete.
- In partial degeneracy, the first two nucleotides are identical but the third (i.e., 3′ base) nucleotide of the degenerate codon differs; for example, CUU and CUC code for leucine.
- Complete degeneracy occurs when any of the 4 bases can take third position and still code for the same amino acid; for example, UCU, UCC, UCA and UCG all code for serine.
- Non-overlapping
- The genetic code is nonoverlapping, i.e.,the adjacent codons do not overlap.
- A nonoverlapping code means that the same letter is not used for two different codons. In other words, no single base can take part in the formation of more than one codon.
- Commaless
- The genetic code is commaless (or comma-free). There is no signal to indicate the end of one codon and the beginning of the next.
- There are no intermediary nucleotides (or commas) between the codons.
- Non-ambiguity
- Non-ambiguous code means that there is no ambiguity about a particular codon.
- A particular codon will always code for the same amino acid.
- While the same amino acid can be coded by more than one codon (the code is degenerate), the same codon shall not code for two or more different amino acids (non-ambiguous).
- Universality
- Universality of the code means that the same sequences of 3 bases encode the same amino acids in all life forms from simple microorganisms to complex, multicelled organisms such as human beings.
- Although the code is based on work conducted on the bacterium Escherichia coli but it is valid for other organisms.
- The genetic code applies to all modern organisms with only minor exceptions, such as the yeast, mitochondria, and the Mycoplasma.
- Polarity
- The genetic code has polarity, that is, the code is always read in a fixed direction, i.e., in the 5′ → 3′ direction.
- It is apparent that if the code is read in opposite direction (i.e., 3′ → 5′), it would specify 2 different proteins, since the codon would have reversed base sequence.
SPECIAL CODONS
A. Chain Initiation Codons
- The triplets AUG and GUG play double roles in E. coli.
- When they occur in between the two ends of a cistron (intermediate position), they code for the amino acids methionine and valine, respectively in an intermediate position in the protein molecule.
- But when they occur immediately after a terminator codon, they act as “chain initiation” (C.I.) signals or “starter codons” for the synthesis of a polypeptide chain.
B. Chain Termination Codons
- The 3 triplets UAA, UAG, UGA do not code for any amino acid.
- When any one of them occurs immediately before the triplet AUG or GUG, it causes the release of the polypeptide chain from the ribosome.
- They are also called as stop codons.
- They are also called chain termination codons because these codons are used by the cell to signal the natural end of translation of a particular peptidyl chain. However, their inclusion in any mRNA results in the abrupt termination of the message at the point of their location even though the polypeptide chain has not been completed.
C. Sense Codons
61 codons, which code for particular amino acids are termed as sense codons.
D. Non-Sense Codons
- Triplets UAA, UAG, UGA do not code for any amino acid.
- They were originally described as non-sense codons.
- However, the so-called non-sense codons have now been found to be of “special sense”.
- These special-sense codons perform the function of punctuating genetic message like a full stop at the end of a sentence.
PATTERNS OF THE GENETIC CODE
- Amino acids with similar structural properties tend to have related codons.
- Aspartic acid codons (GAU, GAC) are similar to glutamic acid codons (GAA, GAG); the difference being exhibited only in the third base (toward 3′ end).
- Similarly, the codons for the aromatic amino acids phenylalanine (UUU, UUC), tyrosine (UAU, UAC) and tryptophan (UGG) all begin with uracil (U).
- All codons with U in the second position specify hydrophobic amino acids (Ile, Leu, Met, Phe, Val).
- All codons with A in the second position specify the charged amino acids, except Arg.
- All the acidic (Asp, Glu) and basic (Arg, Lys) amino acids have A or G as the second base.
READING FRAMES AND OPEN READING FRAMES
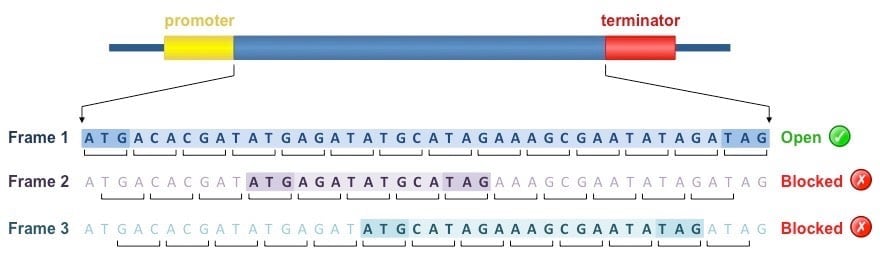
- The mRNA sequence can be read by the ribosome in three possible reading frames.
- Usually only one reading frame codes for a functional protein since the other two reading frames contain multiple termination codons.
- However, in some bacteriophage, overlapping genes occur which use different reading frames.
- An open reading frame (ORF) is a run of codons that starts with ATG and ends with a termination codon, TGA, TAA or TAG.
- Coding regions of genes contain relatively long ORFs unlike non-coding DNA where ORFs are comparatively short.
- The presence of a long open reading frame in a DNA sequence therefore may indicate the presence of a coding region.
- Computer analysis of the ORF can be used to deduce the sequence of the encoded protein.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- http://www.cuchd.in/elibrary/resource_library/University%20Institutes%20of%20Sciences/Fundamentals%20of%20Biochemistry/Chap-30.pdf
- http://www.whsd.net/userfiles/1666/Classes/21126/Genetics%20and%20Heredity%20Completed%20notes.pdf
